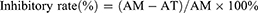Back to Journals » Journal of Inflammation Research » Volume 14
Exogenous Plant gma-miR-159a, Identified by miRNA Library Functional Screening, Ameliorated Hepatic Stellate Cell Activation and Inflammation via Inhibiting GSK-3β-Mediated Pathways
Authors Yu WY , Cai W, Ying HZ, Zhang WY, Zhang HH, Yu CH
Received 4 February 2021
Accepted for publication 22 April 2021
Published 24 May 2021 Volume 2021:14 Pages 2157—2172
DOI https://doi.org/10.2147/JIR.S304828
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Monika Sharma
Wen-Ying Yu,1,* Wei Cai,2,* Hua-Zhong Ying,1 Wen-You Zhang,1 Huan-Huan Zhang,1 Chen-Huan Yu1,3,4
1Zhejiang Key Laboratory of Experimental Animal and Safety Evaluation, Zhejiang Academy of Medical Sciences (Hangzhou Medical College), Hangzhou, People’s Republic of China; 2Department of Traditional Chinese Medicine, Zhejiang Pharmaceutical College, Ningbo, People’s Republic of China; 3Institute of Cancer and Basic Medicine, Chinese Academy of Sciences, Hangzhou, People’s Republic of China; 4Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Hangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chen-Huan Yu
Institute of Cancer and Basic Medicine, Chinese Academy of Sciences, Dongfang Street 150, Hangzhou, Zhejiang, 310018, People’s Republic of China
Email [email protected]
Purpose: Plant-derived exogenous microRNAs (miRNAs) regulate human physiological functions by blocking the translation of target mRNAs. Although several computational approaches have been developed to elucidate the interactions of cross-species miRNAs and their targets in mammals, the number of verified plant miRNAs is still limited, and the biological roles of most exogenous plant miRNAs remain unknown.
Methods: A miRNA mimic library-based phenotypic screening, which contained 8394 plant mature miRNAs published in the official database miRbase, was performed to identify more novel bioactive plant miRNAs for the prevention of hepatic fibrosis. Inhibition of candidates for the activation of hepatic stellate cells (HSCs) and the underlying mechanisms were evaluated in TGF-β 1- and PDGF-exposed HSC models. The protective effects of the candidates against CCl4-induced liver fibrosis were evaluated in a mouse model.
Results: Among the 8394 plant mature miRNAs reported in the official database miRBase, five candidates were found to effectively inhibit the differentiation of HSCs. gma-miR-159a (miR159a) exerted the strongest inhibitory activities on both TGF-β 1- and PDGF-induced HSC activation and proliferation by inhibiting the GSK-3β-mediated NF-κB and TGF-β 1 pathways. Moreover, miR159a was mainly accumulated in the liver after intravenous injection, and it reduced CCl4-induced hepatic fibrosis and inflammation in mice.
Conclusion: Results indicated that miR159a has the therapeutic potential for preventing hepatic fibrosis. This study provides a novel strategy for achieving natural nucleic acid drugs.
Keywords: cross-kingdom regulation, miRNA library, GSK-3β, apoptosis, hepatic fibrosis, inflammation
Introduction
The incidence rate and mortality rate of liver cirrhosis, which is one of the most common cause of deaths worldwide, have rapidly increased in developing countries in recent years. Liver fibrosis is not only a crucial stage but also an important pathological characteristic during the progression from chronic liver injury to cirrhosis.1 Given that this process can be reversed, early intervention and treatment are very important to prevent the occurrence and development of liver disease. Despite the great advancement in the study of the mechanisms of pathogenesis-induced liver fibrosis, no effective antihepatic fibrosis drugs have been approved by US Food and Drug Administration for clinical use. Owing to the lack of evidence-based medical supports, both the American Association for the Study of Liver Diseases and the European Association for the Study of the Liver have released few guidelines or a consensus opinion on liver fibrosis.2,3 Therefore, developing safe and effective hepatoprotective drugs have been the focus of research to prevent the pathogenesis of hepatic fibrosis.
MicroRNAs (miRNAs) are a kind of non-encoded single-chain small molecules composed of 19–25 nucleotides. Many studies have confirmed that miRNAs can recognize target mRNAs to block their translation through base-pairing rules and are involved in various physiological and pathological processes of eukaryotes, such as embryonic differentiation, fertilization, cell adhesion, inflammatory response, and tumor metastasis.4,5 More interestingly, emerging evidence indicates that plant-derived miRNAs, as a novel plant and/or food component, could shuttle across different eukaryotic species and cross-kingdom regulate the mammalian gene expression.6,7 Since Zhang et al8 first discovered the cross-kingdom regulation of exogenous plant miRNAs and found that rice miR168a increases the plasma levels of low density lipoprotein (LDL) in mice by inhibiting LDL receptor adaptor protein 1, additional plant miRNAs, including broccoli miR159, danshen miR-1, and honeysuckle MIR2911, with obvious antitumor and antiviral activities have been identified.9–11 Although the potential scope and source of exogenous miRNAs derived from dietary food and medicinal plants remain controversial,12,13 they had been considered as the 8th nutriment. Recently, several computational approaches have been developed to elucidate the interactions of cross-species miRNAs and their targets in mammals.14,15 However, miRNAs have selective distribution in different tissues, and even the same miRNA can play distinct roles in disease progression.16,17 Thus, the biological roles of most exogenous plant miRNAs remain unknown. In this study, a miRNA mimic library-based functional screening, which contained 8394 plant mature miRNAs published in the official database miRbase (version 22.0), was performed to identify more plant miRNAs with obvious antihepatic fibrotic potentials both in vitro and in vivo. Among all candidates, gma-miR-159a (miR159a), which was derived from soybean, was identified as the top suppressor of TGF-β1- or CCl4-induced hepatic stellate cell activation. These results not only provide a new strategy for discovering a wealth of useful therapeutic agents in plants but also contribute to the study of indigenous remedies with unknown medicinal substances. The process was showed schematically in Figure 1.
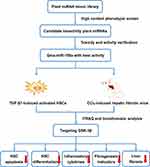 |
Figure 1 The flow chart of the program. |
Materials and Methods
miRNA Mimic Library-Based Functional Screening
The library of chemically synthesized plant miRNA mimics, which contained 8394 plant mature miRNAs reported in the official database miRbase (version 22.0), were obtained from RIBOBIO Biotechnology Company (Guangzhou, China).
The commercially available human hepatic stellate LX-2 cells (HSCs; catalog number: CL-0560) and human normal hepatic LO-2 cells (catalog number: CL-0111) were purchased from Wuhan Procell Company (China). The cell line was authenticated by STR analysis. This study was approved by the ethics committee of Zhejiang Academy of Medical Sciences (Approval No. 2018R1013). The cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 U/mL streptomycin at 37°C in a humidified 5% CO2 incubator.
HSC activation and differentiation were induced by seeding the cells into 384-well plates at a density of 5000 cells/well. The cells were treated with 100 μL of OPTI-MEM culture medium (containing 10 μg/mL TGF-β1 or PDGF-AA) and then incubated at 37°C in a humidified 5% CO2 incubator for 24 h.18–20 The cells were transfected with each different miRNA mimic whose final concentration was 100 nmol/L in each well. A mimic control (100 nmol/L) was used as the negative control, whereas sorafenib (10 μmol/L) was utilized as the positive control. The cells were automatically imaged 0 and 48 h after initiation of miRNA mimic treatment by using the Operetta High Content Screening System (PerkinElmer, USA) in brightfield mode. Five field-of-views were selected in each well by using the Harmony software under the “digital phase contrast” option. The area of each cell was computed using the “calculate morphology properties” option to obtain Figure 1A and B. Inhibitory rate (%) was calculated as follows:
where AM was the average cell area of the model control group, and AT was the average cell area of the miRNA-treated group.
MTT Assay
Inactivated HSCs were seeded in a 96-well plate at a density of 10,000 cells/well, treated with 100 μL of OPTI-MEM culture medium containing 10 μg/mL of TGF-β1, and then incubated at 37 °C in a humidified 5% CO2 incubator for 24 h. The activated HSCs were treated with miR159a (100 nmol/L) or sorafenib (10 μmol/L) as the positive control for 24 h. After incubation, 20 μL of MTT solution (5 mg/mL; Sigma-Aldrich Company, USA) was added to each well, and the cells were incubated at 37 °C for 4 h. When the supernatants were removed, 150 μL of DMSO was added to each well to dissolve the formazan crystals. The optical density of each well was detected at 490 nm by using the Spectra Max M4 microplate reader (MD, USA).
The LO-2 cells were also used to investigate the cytotoxicity of plant miRNAs. The experimental procedure was performed as described above.
RT-PCR Analysis
Total RNA was extracted from the HSC cells by using the Trizol reagent (Thermo Scientific, US) and then reverse-transcribed into complementary DNA (cDNA) by using a cDNA kit (Yeasen Inc., Shanghai, China) following the manufacturer’s instructions. The expression levels of α-SMA, TIMP-1, and Col I were determined using PrimeScript RT-PCR kits (Takara, Shiga, Japan) with GAPDH as the internal reference. Real time-PCR was performed on the ABI7500 quantitative PCR instrument (ABI Company, US). The primers were designed and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) (Table S1). Fold changes of gene expression were calculated via the 2−∆∆Ct method.
Cell Apoptosis Detection
The cells were inoculated in 6-well plates with 2×105 cells each well and then randomly divided into four groups: control group, TGF-β1-treated group, gma-miR-159a (miR159a)-treated group, and mimic control-treated group. The final tested concentrations of miR159a in the experiment were 100 nM. After treatment with 5 μg/mL of TGF-β1 for 24 h, the cells were washed with PBS twice and then mixed with PBS to obtain the suspension (5×105 cells/mL), and then 200 μL of the suspension was mixed with propidium iodide and annexin V-FITC (100 test; Multisciences Biotech, Co., Ltd) for 20 min. The apoptosis of HSCs was detected via flow cytometry. The data were presented as the percentages of annexin V-positive cells in the total cells.
Differentially Expressed Proteins Assessed by iTRAQ Analysis
iTRAQ combined with LC-ESI-MS/MS analysis was employed to investigate the differentially expressed proteins in TGF-β1-treated HSCs after transfection with the miR159a mimic or the mimic control. In brief, 100 μg of the cell lysates was digested with Trypsin Gold reagent (Promega, Madison, USA) at 37 °C for 16 h, and then the peptides were reconstituted in 0.5 M of TEAB and 8-plex iTRAQ reagent (Applied Biosystems, USA). The peptides were marked with isobaric tags at room temperature for 2 h. Data acquisition was performed with a Triple TOF 5600 System (AB SCIEX, ON)by LC-bio Company, Hangzhou, China. Quantitative protein ratios were normalized by the median ratio in Mascot. Only the ratios with p-values < 0.05 and fold changes of >2.0 were considered as statistically significant.
Prediction of miRNA Target Gene
MiRanda, the algorithm of which is based on the removal of Max-Energy target genes more than −10, was used to predict the targets of miR159a. The intersection of miR159a targets generated by the miRanda algorithm and iTRAQ analysis was calculated by Venn diagram tool. The expressions of these potential targets of miR159a were verified by Western blot analysis.
Dual-Luciferase Reporter Assay
The dual-luciferase reporter system was used to investigate whether glycogen synthase kinase-3β (GSK-3β) was the direct target of miR159a in accordance with a previously described method with minor modifications.21 The full-length fragment of wild-type or mutant forms of the GSK-3β gene was cloned into pGL3 vector (Specification: 20 μg; Promega, USA) and transfected into HEK293T cells by using Lipofectamine 3000. Sixteen hours after incubation, dual luciferase activity was measured by a multifunctional microplate reader (MD, USA).
Immunofluorescence Staining
The cells in different groups were grown on 6-well dishes and then treated with 4% formaldehyde (pH 7.2) for 1 h. The permanent cells were washed with normal PBS twice and blocked with 5% bovine serum albumin (BSA) and 0.3% Triton X-100 in PBS for 0.5 h. After treatment, the cells were stained with GSK-3β (Santa Cruz, USA) primary antibodies overnight at 4 °C. The cells were washed with PBS twice and then stained with Texas red-labeled secondary antibodies for 3 h. The cells were washed twice and then stained with 4′,6-diamidino-2-phenylindole (DAPI). Fluorescence images were obtained using a BMI 3000 fluorescence microscope (Zeiss, Germany).
Cytoskeleton Assay
The slides of hepatic fibroblasts were prepared following the aforementioned method and then stained with FITC-labeled phalloidin (KAIJI Biotech Co. Ltd., China) to visualize cytoskeleton, whereas Hoechst 33342 was used to visualize nuclei. F-actin distribution in HSCs was assayed using a BMI 3000 fluorescence microscope (Zeiss, Germany).
Preparation of CCl4-Induced Hepatic Fibrotic Mice and miR159a Treatment
Male ICR mice were bought from Zhejiang Laboratory Animal Center (Hangzhou, China). The mice were fed with filtered water and sterilized food under standard specific pathogen-free conditions. This experiment was approved by the ethics committee of Zhejiang Laboratory Animal Center (Approval No. 2018R1014) and followed the Chinese guidelines for the welfare and treatment of laboratory animals (GB/T 35823–2018).
The mice (4 weeks old) were intraperitoneally injected with 0.1 mL of peanut oil containing 20% CCl4 twice a week for 6 weeks to induce hepatic fibrosis.22,23 The mice with hepatic fibrosis were randomly divided into a model group, a miR159a-treated group, and a silymarin-treated group (as the positive control group). Each group had eight mice. Another eight normal mice (10 weeks old) were used as the control group. Four weeks after CCl4 challenge, the mice in the miR159a-treated group were intraperitoneally injected with 300 nM/kg of miR159a agomir once a week for 2 weeks, whereas the mice in the silymarin-treated group were orally administrated with 150 mg/kg of silymarin once a day for 2 weeks. At the end of the experiment, all mice were sacrificed under anesthesia (isoflurane). Blood samples were collected from celiac artery, and liver samples were weighed and conserved for further analysis.
To investigate the in vivo distribution of miR159a, three mice with hepatic fibrosis were selected and then intraperitoneally injected with 300 nM/kg of miR159a agomir labeled with Cy3 dye, while other 3 model mice were injected with 300 nM/kg of mimic labeled with Cy3 dye. Six mice from model group and normal group were intraperitoneally administrated with saline. About 1 h after intraperitoneal injection, all mice were anaesthetized by isoflurane and the fluorescence intensity of Cy3-labeling miR159a was measured by using Lumina LT animal imaging system (PE company, USA).
Western Blot Analysis
The cells or liver samples were collected and washed with normal saline twice. Total protein was extracted using a protein extraction solution (Servicebio Company, Wuhan, China). Electrophoretic separation was conducted in a protein extraction solution by using 12.5% SDS-PAGE. After separation, the protein was transferred from the separation gel onto a PVDF membrane by electrorotation. The membrane was incubated overnight with the antibodies and with a secondary antibody for 2 h. The protein bands were developed, and their luminescence was detected using the ECL kit.
Histopathological Analysis
A part of the liver sample was put into 10% formalin and then embedded in paraffin. Morphological and fibrotic changes in liver tissues were observed via H&E staining and Masson staining, respectively. Immunohistochemistry analysis was performed to investigate the expression of α-SMA and TGF-β1 (two conventional indicators of hepatic fibrosis) in hepatic tissues. Histopathological features were analyzed in the same manner as in our previous study.22 Histological changes in each liver tissue were individually calculated in three aspects, namely, steatosis, fibrosis, and inflammation, and then appraised by counting the scores of each item.
Fluorescence in situ Hybridization (FISH) Analysis
After prehybridization for 1 h at 37 °C, the mouse liver tissues were treated with a hybridization buffer containing 50 nM of FAM-labeled miR159a probe (synthesized by Bioster, Wuhan, China) at 37 °C overnight. The sections were then blocked with 1% BSA and 3% normal goat serum in PBS for 1 h and then incubated with mouse anti-GSK-3β antibody (Proteintech, USA) overnight. The sections were washed three times with TBS and then incubated with goat anti-mouse IgG for 1 h. The sections were washed three times in PBS, once in DEPC water, and then incubated with DAPI for 20 min to visualize the nuclei. Finally, the sections were washed once with DEPC water and treated with 30% glycerin. Immunofluorescence images were obtained using a confocal microscope (Olympus, Japan).
Culture Medium and Serum Biochemical Marker Analysis
The supernatant was collected after centrifugation at 3000 rpm for 20 min at 4 °C. The inflammatory markers (TNF-α and IL-6) and the fibrotic markers (tissue inhibitor of metalloproteinase 1 [TIMP-1] and type I collagen [Col I]) were determined via ELISA.
About 0.7 mL of the arterial blood was collected and put into the anticoagulated tubes, which were pretreated with EDTA-Na2. Levels of fibrotic indicators (alanine aminotransferase [ALT], aspartate aminotransferase [AST], type III procollagen [PC III], and type IV collagen [Col IV]) in the serum of the mice were measured via ELISA. All ELISA kits were obtained from Boster Biological Technology Co. Ltd. (Wuhan, China).
Statistical Analysis
The data were presented as means±standard deviation (SD) and analyzed via one-way ANOVA by Tukey’s test with SPSS software (ver. 17.0). P values <0.05 indicated statistical significance.
Results
MiR159a Had the Strongest Inhibition on Both TGF-β1- and PDGF-AA-Induced Hepatic Stellate Cell Activation
In this study, TGF-β1 and PDGF were used to promote the proliferation and differentiation of HSCs. Under an inverted microscope, the shape of HSCs with TGF-β1 or PDGF-AA stimulation changed from a small round into a spindle. However, after stimulation with sorafenib (a PDGF blocker) or plant mimics, the shape of HSCs became elliptical or round in varying degrees (Figure 2). On the basis of the experimental results of cell imaging data clustering, candidate plant miRNAs with potent inhibitory activities were screened out by identifying the training sample set and via machine self-learning with a high-content screening system. Among the 8394 plant mature miRNAs reported in the official database miRBase, five candidates were found to effectively inhibit the HSC differentiation, and these candidates had a stronger inhibition than the positive control sorafenib. The results of exogenous plant miRNA library functional screening were analyzed. The inhibitory effects of those five candidate plant miRNAs on HSC proliferation were measured via MTT assay. Results showed that those five candidates could also substantially reduce HSC proliferation. miR159a exhibited the strongest inhibitory activities on both TGF-β1- and PDGF-induced HSC activation and proliferation in vitro. It not only considerably decreased the levels of inflammatory markers (TNF-α and IL-6) and fibrotic markers (TIMP-1 and Col I) in the cell culture mediums but also remarkably reduced the mRNA expressions of α-SMA, TIMP-1, and Col I in the miR159a-treated HSCs compared with those in TGF-β1-activated HSCs. Unlike sorafenib, all those candidate miRNAs did not show any evident cytotoxicity to normal liver LO2 cells, indicating that they were less toxic than sorafenib.
Furthermore, the apoptotic rates of the miR159a-treated group were 10-fold higher than those of the control group, indicating that miR159a induced the apoptosis of TGF-β1-activated HSCs. Therefore, miR159a was considered a potential antihepatofibrotic agent for further research.
GSK-3β Was One of the Targets of miR159a
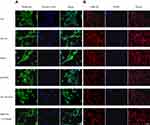
MiRNAs regulate the direction HSC differentiation by regulating many genes via a post-transcriptional protocol. Therefore, iTRAQ-based quantitative proteomic analysis was performed for the global profiling of inactivated HSCs after miR159a transfection. In this study, 86 differentially expressed proteins associated with miR159a overexpression were identified by iTRAQ analysis (Figure 3). Moreover, additional differentially expressed proteins were identified via miRanda bioinformatics predictions, which is a well-known web service for predicting the targets of certain miRNAs. A total of 199 targets were predicted by the miRanda algorithm. On the basis of the degree of downregulation and the presence of a miRNA binding site in the 3ʹ UTR of the target gene, seven candidates were considered in the intersection of the miR159a targets generated by the miRanda algorithm and iTRAQ analysis. However, among those seven targets, the expression levels of GSK-3β, Notch1, and Smad2 were substantially downregulated in the miR159a-treated group and the normal control group compared with those in the model group or the mimic control-treated group (Supplementary Figure S1). Given that GSK-3β, the molecular function of which in the pathogenesis of hepatic fibrosis is well known, was the most differentially expressed gene among the three aforementioned targets, it was considered as the main target of miR159a for further investigation.
 |
Figure 3 GSK-3β was the target of miR159a. (A) Heatmap of fold change of proteins screened out in miR159a-treated HSCs by iTRAQ analysis. (B) Venn diagram showed down-regulated genes in miR159a-treated HSCs. Blue diagram showed targets of miR159a by using miRanda algorithm, while red diagram showed 2-folds down-regulated proteins in miR159a-treated HSCs by iTRAQ analysis. The overlap showed the number of potential targets of miR159a. (C) The expressions of 7 potential targets were verified by Western blot analysis. The relative quantitative results were shown in Supplement Figure S1. (D) Diagram of GSK-3β 3ʹ-UTR-containing reporter constructs (Up). Relative repression of luciferase expression standardized to a transfection control was analyzed by luciferase reporter assays. MiR159a report construct, containing a wild-type or a mutated GSK-3β 3ʹ-UTR, were transfected into H293T cells (down). All values indicated the mean ± SD (n=3). Different letters indicated statistically significant differences, P<0.05 (Tukey’s test). |
Dual-luciferase assay was performed to validate whether GSK-3β is the potential miR159a target. The relative activity of luciferase reporter containing the wild-type 3′ UTR fragment of GSK-3β was considerably reduced, but luciferase activity did not notably change when the binding sites were mutated. These results suggested that miR159a inhibited the expression of GSK-3β in vitro.
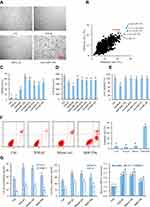
MiR159a Inhibited the Activation of HSCs Induced by TGF-β1
HSC activation is associated with cytoskeleton remodeling of actin, including F-actin upregulation and morphological changes (from round to spindle). As shown in Figure 4, in the control group, the cytoskeleton of the inactivated HSCs had a diffused distribution, the length of F-actin filaments was irregular and uneven, and the shapes of cells were round and oval. However, the fluorescence expression of F-actin in the TGF-β1-induced HSCs was remarkably increased compared with that in the control group. In the TGF-β1-treated group, the F-actin filaments were concentrated along the longitudinal axis of the cells and arranged parallel to the thin strips of stress fibers. After the miR159a or GSK-3β inhibitor AR-A014418 intervention, the F-actin filaments were distributed desultorily, and their green fluorescence intensity was markedly decreased than that in the TGF-β1-treated group. However, all those changes induced by miR159a in the activated cells could be reversed by ADP treatment.
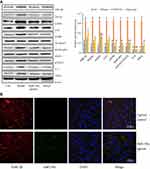
MiR159a Reduced the Expression of GSK-3β and Its Downstream Proteins
After TGF-β1 stimulation, the protein expression levels of GSK-3β and its downstream proteins (mTOR, p-p70S6K, NF-κB, IL-6, TNF-α, p-STAT3, TGF-β1, p-Smad2, Col I, and α-SMA) in the activated HSCs were notably increased compared with those in the control group, whereas miR159a inhibited the increased protein expression levels of GSK-3β and its downstream proteins induced by TGF-β1 in the HSCs (Figure 4 and Supplementary Figure S2). Treatment with the GSK-3β inhibitor AR-A014418 also inhibited TGF-β1-induced activation of the GSK-3β-mediated mTOR/NF-κB and TGF-β1/Smad2 pathways. However, the GSK-3β agonist LY294002 could reverse the inhibition of miR159a on GSK-3β activation induced by TGF-β1 in the HSCs. These results indicated that miR159a inhibited TGF-β1-induced activation of the GSK-3β-mediated pathways by targeting GSK-3β.
MiR159a Improved Hepatic Fibrosis and Inflammation in Mice Induced by CCl4
In the control group, the hepatic surface was smooth with eumorphism in histology. After 6 weeks of CCl4 induction, the liver of the mice showed severe fibrotic symptoms. The hepatic tissues of the model mice became hard, greasy, and coarse with large amounts of white plaques on liver surface. Histopathological examination further confirmed obvious lipid deposition, fibroblast activation, and neutrophil infiltration, as well as increased expression levels of α-SMA and TGF-β1 in the hepatic tissues of the model mice. However, those features could be hardly observed in the normal mice (Figure 5). Administration with miR159a agomir for 2 weeks not only substantially reduced lipid deposition, fibroblast activation, and neutrophil infiltration but also downregulated the expression levels of α-SMA and TGF-β1, whose pathological scores were considerably less than those of silymarin (Supplementary Figure S3), indicating that miR159a has a good therapeutic potential on CCl4-hepatic fibrosis in mice.
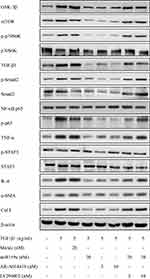 |
Figure 5 MiR159a reduced the expressions of GSK-3β and its downstream induced by TGF-β1 in HSCs. Both miR159a and GSK-3β inhibitor AR-A014418 down-regulated the expressions of GSK-3β and its downstream (mTOR, p-p70S6K, p-Smad2, NF-κB and p-STAT3). However, GSK-3β agonist LY294002 could reverse the inhibition of miR159a on the activation of GSK-3β induced by TGF-β1 in HSCs. The relative quantitative results were shown in.Supplement Figure S2. |
By contrast, the hepatic indexes, as well as the serum hepatic function biomarkers (ALT, AST, PC III and C IV) and inflammatory cytokines (TNF-α, IL-1β andIL-6), of the model mice were considerably increased compared with those of the normal mice (Table 1). However, administration with miR159a markedly reduced all those biomarkers, indicating the antihepatic fibrotic and anti-inflammatory effects of miR159a.
 |
Table 1 Effects of miR159a Agomir on Hepatic Index and Serum Biomarkers in CCl4-Induced Hepatic Fibrosis Mice |
MiR159a Was Mainly Distributed in the Hepatic Tissues of Mice
About 15 min after intraperitoneal injection with 20 nM of Cy5-labeled miR159a agomir, the fluorescence images of mice were obtained using an animal imaging system. As shown in Figure 6, only the chests of the miR159a-treated mice presented strong fluorescence signals, whereas no fluorescence signals were observed in the other groups. This result indicated that miR159a was mainly distributed in the hepatic tissues of mice.
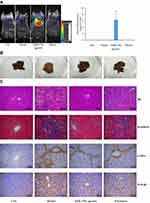 |
Figure 6 MiR159a ameliorated CCl4-induced hepatic fibrosis in mice. (A) Imaging in vivo and the relative quantitative results of miR159a agomir labeled with Cy3 dye in mice. (B) The histologic appearance of fibrotic changes in hepatic tissues. (C) The histopathological changes in hepatic tissues stained by HE and MASSON. The expression of α-SMA and TGF-β1 in hepatic tissues detected by immunohistochemistry. The pathological scores of each group as well as the relative expressions of α-SMA and TGF-β1 were showed in Supplement Figure S3. Scale bars, 100 μm. |
The expression levels of GSK-3β and its downstream proteins (mTOR, NF-κB, IL-6, TNF-α, p-STAT3, TGF-β1, Col I, and α-SMA) in the model group were substantially increased compared with those in the control group, whereas those in the miR159a- or silymarin-treated group were considerably decreased compared with those in the model group (Figure 7). Furthermore, FISH analysis revealed that miR159a labeled with FAM could be detected in the hepatic tissues of the miR159a-treated mice, whose localization was mainly in the intercellular substances and cytoplasm. However, it was hardly detected in the control group. These results indicated that miR159a could be took up by mouse hepatic cells and subsequently downregulate the expression levels of key proteins of GSK-3β-mediated NF-κB and TGF-β1 in the liver tissues of the mice.
Discussion
Numerous studies recently demonstrated that plant miRNAs, as novel dietary functional components, can be absorbed by the gastrointestinal cells via the SIDT1 pathway, thereby entering the bloodstream and regulating the expression of endogenous mRNAs.8 Owing to the 2-O-methylation of the terminal region of these genes, the structures of some plant miRNAs cannot be destroyed after being treated with strong oxidants (such as NaIO4), strong corrosive agents (such as formalin and toluene), and even gastrointestinal digestion and decomposition; its structure remained intact and could be detected by RT-PCR, indicating that the structures of plant miRNAs are highly stable.24 Given that there are no exactly the same gene sequences between plants and animals, the plant miRNAs found in animal serum or organ tissue must have come from plants and not from animal endogenous miRNAs or animal endogenous RNA fragments.25 Therefore, plant miRNAs could be potentially used as therapeutic and dietary supplements against human diseases.
miRNA libraries for loss or gain of function are widely used to identify new biological mechanisms of miRNA, including cell proliferation, differentiation, apoptosis, and survival.26 miRNA mimics synthesized by chemical methods can elevate the expression of mature exogenous or endogenous miRNA in cells and be useful in screening for gain of functions.27,28 In this study, additional plant miRNAs exerting therapeutic potential were identified via high-throughput phenotypic screening for antihepatic fibrotic miRNAs by using a library of chemically synthesized plant miRNA mimics. Visible morphological differences between normal HSCs and activated HSCs were identified by a machine learning algorithm to establish a rapid and accurate method for screening potential plant miRNAs. Among the 8394 plant mature miRNAs reported in the official database miRBase, five miRNAs, namely, gma-miR-159a, csi-miR-482b, crt-miR-168, zma-miR-393a, and cca-miR-167, were found to have obvious inhibitory activities on both TGF-β1- and PDFG-AA-induced HSC activation in vitro. Further studies showed that miR159a remarkably inhibited HSC activation and proliferation induced by TGF-β1 compared with the other four candidates. More importantly, miR159a did not affect the viability of normal liver cells, indicating the potential of miR159a for treating hepatic fibrosis.
miRNAs regulate the direction of HSC differentiation by wielding a post-transcriptional protocol on large numbers of target genes.29 A combined analysis of the results of iTRAQ-based proteomic identification and miRanda algorithm found seven candidate targets of miR159a. Western blot analysis revealed that after TGF-β1 challenge, the expression of GSK-3β was substantially increased in the activated HSCs, which could be reversed by miR159a. Dual-luciferase reporter analysis also showed that miR159a markedly inhibited the luciferase activities of H293T cells transfected with the miR159a seed binding site of GSK-3β, but this effect disappeared when the nucleotides of the binding site were mutated. These results indicated that GSK-3β is one of the targets of miR159a. Previous studies demonstrated that GSK-3β is an evolutionarily conserved serine/threonine kinase widely found in mammalian eukaryotic cells. Aside from regulating the activity of glycogen synthase, GSK-3β not only acts as an oncogene but also participates in various cellular processes, including cell cycle control, DNA damage and repair, gene transcription, and cell apoptosis.30,31 GSK-3β is associated with the development of numerous human diseases, including Type 2 diabetes, atherosclerosis, cardiomyopathy, fibrosis, aging, infertility, and certain cancers.31,32 Remarkably, GSK-3β activation is involved in the development of human liver cirrhosis, whereas GSK-3β inhibition prevents fibroblast activation and fibrogenesis, resulting in decreased levels of ROS and subsequently inactivating various downstream factors, including mTOR, NF-κB, and TGF-β1 signaling, as well as JAK/STAT3 phosphorylation.33 Therefore, selective targeting of GSK-3β presents a promising strategy for ameliorating several pathological conditions of liver fibrotic diseases. In this study, treatment with miR159a or the GSK-3β inhibitor AR-A014418 reduced the increased expression levels of GSK-3β and its downstream proteins, as well as those of activated cell phenotypes induced by TGF-β1, which were abolished by the GSK-3β agonist LY294002. These results indicated that miR159a inhibits TGF-β1-induced HSC activation by inactivating GSK-3β signaling.
The reliability of in vitro data was verified. The in vivo distribution and therapeutic efficacy of miR159a were investigated on the chemically induced liver fibrosis and inflammation in vivo. After 6-week exposure to CCl4, obvious mesenchymal alterations appeared in the liver tissues, including thickening of the basal lamina, lipid accumulation, fibrous hyperplasia, and inflammatory cell infiltration. Moreover, the serum levels of hepatic functional markers (ALT, AST, PC III, and C IV) and inflammatory cytokines (TNF-α, IL-1β, and IL-6) were substantially increased in the mice. However, treatment with 300 nM of miR159a agomir once a week for 2 weeks evidently ameliorated CCl4-induced lipid accumulation and liver fibrosis; reduced the increased expression levels of ALT, AST, PC III, C IV, TNF-α, IL-1β, and IL-6 in serum; and suppressed the elevated expressions of GSK-3β and its downstream key proteins of the NF-κB and TGF-β1 pathways in hepatic tissues. These results indicated that miR159a could be used as a potential agent for hepatic fibrosis treatment. Moreover, animal imaging results showed that agomir was mainly expressed in the liver tissues of the mice, implying that it has a specific distribution pattern in tissues. FISH analysis further revealed that the positive expression of miR159a could be clearly visualized in the intercellular substances and cytoplasm. Thus, we conjectured that miR159a could be absorbed by mouse hepatic cells and then downregulate the expression levels of GSK-3β-mediated signaling pathways in liver tissues.
In conclusion, the antifibrotic activities of soybean-derived miR159a was first identifies by exogenous plant miRNA library functional screening analysis. MiR159a not only induced apoptosis and inactivation of HSCs in vitro through suppressing GSK-3β-mediating NF-κB and TGF-β1 pathways, but also ameliorated the liver fibrosis and inflammation in CCl4-exposed mice. Therefore, miR159a could be used as an effective therapeutic agent for prevention of hepatic fibrosis. This study also provided a novel strategy for discovering natural nucleic acid-like candidates in the plant medicine.
Abbreviations
ALT, alanine aminotransferase; AST, aspartate aminotransferase; BSA, bovine serum albumin; CCl4, carbon tetrachloride; Col IV, type IV collagen; DAPI, 4′,6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridization; GSK-3β, glycogen synthase kinase-3β; HSCs, hepatic stellate cells; LDL, low density lipoprotein; miRNAs, microRNAs; mTOR, mammalian target of rapamycin; NF-κB, nuclear factor kappa-B; PC III, type III procollagen; PDGF, Platelet derived growth factor; STAT3, signal transducer and activator of transcription 3; TGF-β1, transforming growth factor β1.
Acknowledgments
This work was supported by Natural Science Foundation of Zhejiang Province (No. LZ17H280001), National Natural Science Foundation of China (No. 81473339 and 81573591), National Natural Science Foundation of Ningbo (No. 2015A610282), and Zhejiang Medical science and technology program (No. 2015ZB104 and 2015KYA065).
Disclosure
The authors declare no conflicts of interest related to this work.
References
1. Wells RG. Hepatic fibrosis in children and adults. Clin Liver Dis. 2018;9:99–101. doi:10.1002/cld.623
2. Wattacheril J, Issa D, Sanyal A. Nonalcoholic Steatohepatitis (NASH) and hepatic fibrosis: emerging therapies. Annu Rev Pharmacol Toxicol. 2018;58:649–662. doi:10.1146/annurev-pharmtox-010617-052545
3. Ying HZ, Chen Q, Zhang WY, et al. PDGF signaling pathway in hepatic fibrosis pathogenesis and therapeutics (Review). Mol Med Rep. 2017;16:7879–7889. doi:10.3892/mmr.2017.7641
4. Matsuyama H, Suzuki HI. Systems and synthetic microRNA biology: from biogenesis to disease pathogenesis. Int J Mol Sci. 2019;21:132. doi:10.3390/ijms21010132
5. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465. doi:10.1002/jcp.27486
6. Zeng J, Gupta VK, Jiang Y, Yang B, Gong L, Zhu H. Cross-kingdom small RNAs among animals, plants and microbes. Cells. 2019;8:371. doi:10.3390/cells8040371
7. Zhang L, Chen T, Yin Y, Zhang CY, Zhang YL. Dietary microRNA-a novel functional component of food. Adv Nutr. 2019;10(4):711–721. doi:10.1093/advances/nmy127
8. Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22(1):107–126. doi:10.1038/cr.2011.158
9. Chin AR, Fong MY, Somlo G, et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. doi:10.1038/cr.2016.13
10. Qin Y, Zheng B, Yang GS, et al. Salvia miltiorrhiza-derived Sal-miR-58 induces autophagy and attenuates inflammation in vascular smooth muscle cells. Mol Ther Nucleic Acids. 2020;21:492–511. doi:10.1016/j.omtn.2020.06.015
11. Zhou LK, Zhou Z, Jiang XM, et al. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020;6:54. doi:10.1038/s41421-020-00197-3
12. Lang C, Karunairetnam S, Lo KR, et al. Common variants of the plant microRNA-168a exhibit differing silencing efficacy for human low-density lipoprotein receptor adaptor protein 1 (LDLRAP1). MicroRNA. 2019;8:166–170. doi:10.2174/2211536608666181203103233
13. Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013;31:965–967. doi:10.1038/nbt.2737
14. Pirrò S, Minutolo A, Galgani A, Potestà M, Colizzi V, Montesano C. Bioinformatics prediction and experimental validation of microRNAs involved in cross-kingdom interaction. J Comput Biol. 2016;23(12):976–989. doi:10.1089/cmb.2016.0059
15. Yu D, Lu J, Shao W, et al. MepmiRDB: a medicinal plant microRNA database. Database (Oxford). 2019;2019:baz070. doi:10.1093/database/baz070
16. Panni S, Lovering RC, Porras P, Orchard S. Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194417. doi:10.1016/j.bbagrm.2019.194417
17. Wang W, Liu D, Zhang X, Chen D, Cheng Y, Shen F. Plant microRNAs in cross-kingdom regulation of gene expression. Int J Mol Sci. 2018;19:2007. doi:10.3390/ijms19072007
18. Wang R, Zhang D, Tang D, et al. Amygdalin inhibits TGFbeta1-induced activation of hepatic stellate cells (HSCs) in vitro and CCl(4)-induced hepatic fibrosis in rats in vivo. Int Immunopharmacol. 2021;90:107151. doi:10.1016/j.intimp.2020.107151
19. Zhang SL, Ma L, Zhao J, et al. The phenylethanol glycoside liposome inhibits PDGF-induced HSC activation via regulation of the FAK/PI3K/Akt signaling pathway. Molecules. 2019;24(18):3282. doi:10.3390/molecules24183282
20. Wang YH, Suk FM, Liu CL, et al. Antifibrotic effects of a barbituric acid derivative on liver fibrosis by blocking the NF-kappaB signaling pathway in hepatic stellate cells. Front Pharmacol. 2020;11:388. doi:10.3389/fphar.2020.00388
21. Li J, Chen W, Yi Y, Tong Q. miR-219-5p inhibits tau phosphorylation by targeting TTBK1 and GSK-3β in Alzheimer’s disease. J Cell Biochem. 2019;120:9936–9946. doi:10.1002/jcb.28276
22. Hu YX, Yu CH, Wu F, et al. Antihepatofibrotic effects of aqueous extract of Prunella vulgaris on carbon tetrachloride-induced hepatic fibrosis in rats. Planta Med. 2016;82:97–105. doi:10.1055/s-0035-1558112
23. Sha M, Gao Y, Deng C, et al. Therapeutic effects of AdipoRon on liver inflammation and fibrosis induced by CCl(4) in mice. Int Immunopharmacol. 2020;79:106157. doi:10.1016/j.intimp.2019.106157
24. Zempleni J, Baier SR, Howard KM, Cui J. Gene regulation by dietary microRNAs. Can J Physiol Pharmacol. 2015;93:1097–1102. doi:10.1139/cjpp-2014-0392
25. Sanchita TR, Asif MH, Trivedi PK, Trivedi PK. Dietary plant miRNAs as an augmented therapy: cross-kingdom gene regulation. RNA Biol. 2018;15:1433–1439. doi:10.1080/15476286.2018.1551693
26. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi:10.1038/nrd.2016.246
27. Palumbo T, Poultsides GA, Kouraklis G, et al. A functional microRNA library screen reveals miR-410 as a novel anti-apoptotic regulator of cholangiocarcinoma. BMC Cancer. 2016;16:353. doi:10.1186/s12885-016-2384-0
28. Ujihira T, Ikeda K, Suzuki T, et al. MicroRNA-574-3p, identified by microRNA library-based functional screening, modulates tamoxifen response in breast cancer. Sci Rep. 2015;5:7641. doi:10.1038/srep07641
29. Zhao Z, Lin CY, Cheng K. siRNA- and miRNA-based therapeutics for liver fibrosis. Transl Res. 2019;214:17–29. doi:10.1016/j.trsl.2019.07.007
30. Zheng H, Yang Z, Xin Z, et al. Glycogen synthase kinase-3beta: a promising candidate in the fight against fibrosis. Theranostics. 2020;10:11737–11753. doi:10.7150/thno.47717
31. Lin J, Song T, Li C, Mao W. GSK-3β in DNA repair, apoptosis, and resistance of chemotherapy, radiotherapy of cancer. Biochim Biophys Acta Mol Cell Res. 2020;1867:118659. doi:10.1016/j.bbamcr.2020.118659
32. Lal H, Ahmad F, Woodgett J, Force T. The GSK-3 family as therapeutic target for myocardial diseases. Circ Res. 2015;116:138–149. doi:10.1161/CIRCRESAHA.116.303613
33. Guo Y, Gupte M, Umbarkar P, et al. Entanglement of GSK-3β, β-catenin and TGF-β1 signaling network to regulate myocardial fibrosis. J Mol Cell Cardiol. 2017;110:109–120. doi:10.1016/j.yjmcc.2017.07.011
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.