Back to Journals » Infection and Drug Resistance » Volume 15
Evidence of Sharing of Carbapenem-Resistant Klebsiella pneumoniae Strains Between Intensive Care Unit Patients and the Environment
Authors Kang J , Li G, Ma M, Lan M, Kang Y, Yang N, Jia W , Zhao Z
Received 6 October 2022
Accepted for publication 10 December 2022
Published 30 December 2022 Volume 2022:15 Pages 7831—7839
DOI https://doi.org/10.2147/IDR.S388085
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Jia Kang,1,* Guangqi Li,1,* Miao Ma,2 Min Lan,3 Yuting Kang,4 Ningai Yang,4 Wei Jia,1,4 Zhijun Zhao1,4
1Medical Experimental Center, General Hospital of Ningxia Medical University, Yinchuan, People’s Republic of China; 2Clinical Laboratory Center, Shaanxi Provincial People’s Hospital, Xi’an, People’s Republic of China; 3School of Clinical Medicine, Ningxia Medical University, Yinchuan, People’s Republic of China; 4Ningxia Key Laboratory of Clinical and Pathogenic Microbiology, General Hospital of Ningxia Medical University, Yinchuan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Jia; Zhijun Zhao, Medical Experimental Center, General Hospital of Ningxia Medical University, Yinchuan, 750001, People’s Republic of China, Tel +86 951-6743543, Email [email protected]; [email protected]
Purpose: Carbapenem-resistant Klebsiella pneumoniae (CR-KP) has emerged as an important public health threat. Intestinal colonization with CR-KP increases the risk of infection and death, especially in intensive care unit patients. To clarify the source of colonizing bacteria is very important to prevent the spread of CR-KP, so the purpose of this study was to explore the relationship between the ward environment and intestinal colonization of CR-KP.
Methods: In this study, 353 environmental swabs from ICU (Intensive Care Unit) wards and 241 anal swab samples from ICU patients were collected and screened on MacConkey plates containing 2 μg/mL ertapenem, and the origin and genotype of CR-KP were analyzed by PCR and sequencing. The sequence type of the strains was also obtained by multi-locus sequence type (MLST) analysis, and plasmid conjugation test was used to clarify whether CR-KP can promote the transmission of drug resistance genes through plasmid integration and rearrangement.
Results: A total of 20 CR-KP environmental strains and 7 intestinal strains were obtained, most of which were blaOXA-48 resistant genotypes. Four different STs were identified by multi-locus sequence type (MLST) analysis, among which the large logarithm was ST485 type, and PFGE clustering showed that the similarity between them was > 85%. In the plasmid transcoupling assay, we report that one of the Klebsiella pneumoniae drug-resistant plasmids was successfully transferred to E. coli, indicating that it may promote the spread of drug-resistant genes through plasmid integration and rearrangement.
Conclusion: Our research suggests that the environment may be a potential source of CR-KP and that there is a need for us to adopt more effective disinfection measures.
Keywords: carbapenem-resistant Klebsiella, intestinal, colonization, environment, ICU, homology
Introduction
Klebsiella pneumoniae has long been recognized as an agent of disease since it was first described by Carl Friedländer as the cause of pneumonia in 1882, and remains among the world’s most common nosocomial gram-negative pathogens.1 It can cause severe pneumonia, bloodstream infections and urinary tract infections2 in patients in the ICU (Intensive Care Unit). Klebsiella pneumoniae can carry a variety of drug resistance genes and can be resistant to ß-lactams, tetracyclines3 and quinolones.4 For a long time carbapenems were considered to be an effective treatment for multi-drug resistant Klebsiella pneumoniae. However, the widespread use of antimicrobial drugs has led to the production of an increasing number of extended spectrum-ß-lactamases (ESBLs) and metallo-ß-lactamases (MBLs),5 which are thus resistant to carbapenems. Carbapenem-resistant Klebsiella pneumoniae (CR-KP) has few treatment options and is a serious public health threat.6
Intestinal colonization of CR-KP is an independent risk factor for systemic CR-KP infection, and colonization precedes or coincides with infection.7 Studies have shown that CRE colonization more than triples the risk of CRE infection in ICU patients, with a concomitant increase in the risk of death from infection.8 In the ICU ward, the mobility of personnel is high, most patients are critically ill and immunocompromised, and there are many invasive treatment procedures, which leads to a greater possibility of bacterial transmission, including through fecal contamination. Asymptomatic colonized patients and healthy medical staff can serve as important reservoirs of infectious agents. And multiple studies have shown that the spread of bacteria can occur between patients, medical staff, medical equipment and clinical settings.9 Previous studies have focused on CR-KP molecular epidemiology and resistance mechanisms,10,11 and have reported how biofilms in the environment exert a high degree of resistance to antimicrobial agents.12 To the best of our knowledge, no studies on environmental-human gut sharing have been performed on CR-KP. However, this information is critical for better understanding the epidemiology of this important pathogen. Therefore, in this study, we screened the ICU environment and the intestinal colonization of CR-KP in patients, and studied its molecular characteristics, aiming to provide some clues about the transmission route of CR-KP.
Materials and Methods
Reagents
Imipenem, meropenem and ertapenem was purchased from Pfifizer. Trypticase Soy Broth (TSB), Mueller Hinton and MacConkey agar were purchased from OXOID. Primer synthesis was performed by Sangon Biotech Shanghai Co. Ltd. Premix Taq and Xba I restriction enzymes were purchased from Takara Biotech Dalian Co. Ltd.
Sample Collection, Bacterial Identification, and Reference Strains
All fecal and anal swab samples were collected from the General Hospital of Ningxia Medical University, a large hospital located in the northwestern city of Yinchuan, China, and were collected non-invasively after informed, written consent was obtained. Reference strain isolates (E. coli ATCC25922, Klebsiella pneumoniae ATCC BAA-1705, and Klebsiella pneumoniae ATCC BAA-1706) and bacterial identification isolates were also obtained from the same hospital. The study population included all patients in the ICU and RICU (Respiratory Intensive Care Unit). Prior to inclusion, all human participants were informed about the main objectives of the study and asked to sign a consent form. The first anal swab was taken within 2 hours of the patient’s admission to the ICU and daily thereafter until CR-KP was detected or the patient left the ICU. If CR-KP was not detected in the stool or anal swab on the first collection but was detected on a subsequent occasion, the participant was considered a CR-KP coloniser and was included in the study. The patient’s age and gender are not limited. Before the disinfection operation of indoor items at 2:00 p. m., environmental bacteria were collected with a physiological saline cotton swab, and then the swab was placed in 3 mL of TSB broth containing a piece of meropenem drug sensitive paper (10μg), and cultured overnight. Patients and surface swabs from ICUs were collected between June and December 2020, and then cultured on MacConkey plates containing 2 μg/mL ertapenem. Species identification and initial sensitivity analysis were performed using MALDI-TOF MS and Vitek2 compact from Biomerieux, France.
Identification of Klebsiella pneumoniae by PCR and Sequencing
Genomic DNA was obtained by boiling method, and then used as a template to detect 16S rRNA gene segment (1465 bp) by PCR. The sequences of primers:27F:AGAGTTTGATCMTGGCTCAG, and 1492R:TACGGYTACCTTGTT ACGACTT. The PCR programs were as follows: denaturation at 94°C 5 min, followed by 30 cycles of 94°C 30s,55°C 40s, and 72°C 1 min, and extension at 72°C 5 min. The PCR products were sequenced by Sangon Biotech Shanghai Co. Ltd.
Drug Susceptibility Test and Detection of Resistance Genes
CR-KP was determined by measuring the minimum inhibitory concentrations of isolates for imipenem and meropenem by the agar dilution method, and the results were interpreted according to the Clinical Laboratory Standards Institute (CLSI) guidelines (M100, 30th). The CR-KP strains were screened by PCR for the presence of the following Drug-resistant genes:blaTEM, blaSHV, blaKPC, blaNDM-1, blaIMP, blaVIM and blaOXA-48. After preparing a 50 µL reaction system, proceed as follows: denaturation at 94 °C for 5 min, followed by 30 cycles of 94°C 30s,55°C~57°C 40s, and 72°C 45s, and extension at 72°C 5 min. The primer sequences, product lengths and annealing temperatures for drug resistance genes are shown in Table 1. Positive amplification products were sequenced and the sequencing results were compared by BLAST.
 |
Table 1 Primer Sequence, Product Length and Annealing Temperature of β-Lactamase Genes |
Genetic Homogeneity Analysis
To determine the genetic homology of CR-KP, pulsed-field gel electrophoresis (PFGE) methods and multilocus sequence typing (MLST) were used. The primers used for MLST and the PCR reaction conditions were referred to the literature.13 Bacterial DNA embedded in small plastic blocks was treated with proteinase K and cleaved by the restriction enzyme XbaI at 37°C for 2.5h. DNA digested by xbai was electrophoresed at 6V for 18.5h, the pulse angle was 120◦, and the pulse time was 6.8 to 35.4 seconds.14 PFGE was repeated three times. K. pneumoniae strains with unique pulse types from environment or human were further typed by MLST according to the published consensus MLST scheme (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). The PFGE and MLST results were analyzed by BioNumerics to construct a similarity tree.
Plasmid Conjugation Test
The conjugation test was performed by the broth mating method using CR-KP (environmental source) as the donor strain and E coli. J53 as the recipient strain. First, take 500 μL each of the donor bacteria and acceptor bacteria in the exponential growth phase and add them to 4 mL of LB broth, and let stand overnight in a 35°C incubator. Then, on the MH solid medium of 2μg/mL meropenem and 200 μg/mL NaN3, the cells were cultured overnight in a 35°C incubator. In this process, the donor and recipient strains were used as negative controls. Finally, combined with the 16S rDNA sequencing results and the agar dilution method to determine the MIC values of imipenem and meropenem, the conjugation strains were identified.15 The experiment was repeated three times.
Results
Source and Genotype of Carbapenem-Resistant Klebsiella pneumoniae
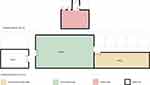
A total of 353 environmental swabs and 241 patients were CR-kp screened at admission to the ICUs from from June to December 2020. 7(2.9%) were CR-kp colonized:5 strains in Ward A of ICU and 2 in RICU. At the same time, we screened 20 strains of CR-KP from the environment. The distribution of these environmental bacteria in the ward is shown in Figure 1, and most of them (18/20) originated from the ward A. Of the environmental CR-KP isolates, 7 were collected from stethoscopes, 4 from bed rail,4 from the computer and 2 from the nurse station. There was only one strain from the ventilator and rack. In terms of time, these environmental bacteria were isolated from September and October, which is close to the isolation time of CR-KP colonized in the patient’s intestine. In addition, 2 intestinal colonizing CR-KP strains were isolated from the respiratory intensive care unit in July. As shown in Table 2, all strains carried β-lactamases gene SHV, and 25 strains carried TEM. Carbapenem resistance genes were mainly blaOXA-48 (21/27), followed by metalloenzymes (blaIMP, 4/27). Two of the 27 strains were found to be New Delhi metallo-lactamase-1 (blaNDM-1). KPC and VIM genes were not detected.
 |
Table 2 Screening Results and β-Lactamase Gene of Carbapenem-Resistant Klebsiella pneumoniae |
Molecular Typing
The molecular profiles of all isolates were determined by pulsed field gel electrophoresis (PFGE), and the sequence profiles of the strains were obtained by multi-site sequence typing (MLST) analysis, both of which were used to characterize the molecular types at the same time. The dendrogram of Figure 2 was generated based on PFGE analysis without weighting the ST included in the graph. Based on a cutoff of 80% genetic similarity, these isolates were grouped into A (isolate No: 229), B (isolate No: 97), C (isolate No: 225, 203, 237, 279, 243, 207, 132, 161, 165, 141, 147, 236, 240, 333, 340, 343, 339, 344, 345, 335, 336, 337, 338, 223), and D (isolate No: 102) clusters (Figure 2). A total of 4 distinct STs were identified among 27 CR-KP. As depicted in Figure 2, ST485 was the most prevalent ST (85.19%). There was one strain each of ST101, ST1737 and ST1905. One strain could not find the corresponding ST type. The patient strains that are clonal to the strains in the environment are 4 out of 7.
 |
Figure 2 Dendrogram showing pulsed-field gel electrophoresis (PFGE) analysis of the 27 carbapenem-resistant Klebsiella pneumoniae isolates. |
Plasmid Conjugation Test
The plasmid transconjugant test was performed for 20 CR-KP isolates from environment, and one Klebsiella pneumoniae resistant plasmid was successfully transferred to E. coli J53. The MIC of the transconjugant for imipenem and meropenem was measured by the agar dilution method (Table 3).
 |
Table 3 Minimum Inhibitory Concentrations (MIC) for Imipenem and Meropenem of the Carbapenem-Resistant Klebsiella pneumoniae Isolate and Its Transconjugant |
Discussion
The colonization of Klebsiella pneumoniae may increase the incidence of corresponding Klebsiella pneumoniae infection in critically ill patients in the ICU,16 which can further markedly prolong the hospital length of stay, increase the medical cost, and the most important hazard is causing high mortality.17 With the widespread colonization of CR-KP and the wide spread of associated resistant genes around the world,18 it has become urgent to investigate the molecular characteristics and colonization pathway of carbapenem-resistant Klebsiella pneumoniae, which may provide a solid basis for an effective control of carbapenem-resistant Klebsiella pneumoniae. To our knowledge, this is the first report of a Klebsiella pneumoniae clonal lineage between ICU ward patients and close contact environments.
In recent years, the research on microorganisms in the ICU medical environment has not stopped. Recent studies have found that the surface of the ICU environment is a repository for fungi,19 and Durán-ManuelEM has found that Acinetobacter baumannii can be clonally spread between medical facilities in the ICU by analyzing 16S rRNA.20 Even CR-KPs in public settings are reported from time to time,21 and the potential transmission of this CR-KP in the environment is worrisome. In this study, 20 strains of CR-KP were screened out from 353 ICU environmental samples, with a detection rate of 5.67% (20/353), including 18 strains in ward A, 2 strains in ward B and no strains in ward C. Comparing the results of intestinal colonization CRE screening, it was found that the high detection rate in ward A may be related to hospitalized patients, because intestinal colonization CRE was screened successively in two patients in two beds in ward A of ICU ward on September 28 and October 10, and CREs were successively detected from the environment in the days before and after this date. Geographically, ward B is connected toward A, while ward C is relatively independent, which may also explain the difference in the detection rate of CR-KP among the three to a certain extent (Figure 1). It is worth noting that several studies22 have shown that handwashing sinks is the source of transmission of CR-KP in the ICU.23 In this study, the detection rate of stethoscope, bed guard and computer CRE was significantly higher than that of other sampling sites, which suggested that we should focus on strengthening the disinfection measures in these parts to reduce its potential for nosocomial transmission. Implementing a more effective cleaning and disinfection programme may be necessary.
ST11 belongs to the dominant clone in Asia. In the molecular epidemiological study of carbapenem-resistant Klebsiella pneumoniae in China, most of the isolates also belong to ST11 and ST15.24,25 Therefore, it was surprising that K. pneumoniae from the homology analysis between the ward environment and the CR-KP colonized in the patient’s intestine, we found that 85.19% (24/27) of the CR-KP belonged to the ST485 type, and PFGE clustering showed that the similarity between them was >85% (Figure 2). These highly homologous strains were all isolated from ward A and B of the comprehensive ICU, and the similarity between the two CR-KP strains from respiratory ICU patients was less than 60%. These results suggest that CR-KPs propagate each other in the environment, especially in close proximity. There is a high degree of homology between the environment and the gut-colonizing CR-KP of the patient in it, which provides strong evidence for the transmission of CR-KP between the patient’s gut and the environment. Numerous studies have shown that admission to the ICU is a risk factor for CR-KP infection, and a review of the clinical cases of seven positive patients revealed that two were subsequently infected with CR-KP, which somewhat confirms this conclusion. Therefore, in order to detect and isolate a “superbug” such as CR-KP early, it is necessary to actively screen high-risk patients admitted to the ICU for CRE.
There was a significant correlation between the type and frequency of drug resistance genes in Klebsiella pneumoniae and their virulence.26 There are differences in the risk of infection caused by different drug-resistant genotypes of CR-KP, NDM-KP was associated with increased risk of BSI compared with KPC-KP.27 Genomic studies of MBLs have shown that NDM and VIM are expressed through genes located on mobile genomic elements and plasmids,5 and that the rich bacterial environment of the gut provides strong conditions for the horizontal transmission of these drug-resistant genes. In this study two strains of NDM-type CR-KP were identified in respiratory ICU patients, but fortunately VIM was not detected. In China, KPC-2, NDM, and OXA-48-like carbapenemases were predominant among CRE clinical isolates.28 However, in this study, OXA-48 carbapenem-resistant Klebsiella pneumoniae dominated (21/27). In order to explore the possible transmission route of the drug resistance gene, we carried out the plasmid conjugation transfer test. In our experiment, one of the 20 environmental CR-KP strains successfully transferred the drug resistance gene to E. coli J53 (Table 3). Chen’s study also highlights how plasmid integration and rearrangements can contribute to the spread of -like genes.29 Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr has been previously detected in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae.30 These results provide important clues for clinical prevention of the spread of carbapenemase-resistant Klebsiella pneumoniae strains.
Conclusion
The results of this study suggest that there is a high similarity between CR-KP colonisation in the ICU environment and in the patient’s gut, and that the environment may be a potential source of intestinal CR-KP colonisation, suggesting the need for more effective cleaning measures to eliminate such a potential problem. However, it remains to be seen whether it is only the CRE in the gut that contaminates the environment or whether it is the environmental CRE that somehow colonises the patient’s gut, or whether it is the bacteria from other patients that pass through the environment and colonise the gut of people in the same ward.
Ethics Approval
The research protocol has been approved by the Medical Research Ethics Review Committee of the General Hospital of Ningxia Medical University (No. 2019-105).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No.81960386), and Key Research and Development Project of Ningxia Hui Autonomous Region (No. 2021BEG03090).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pendleton Jack N, Gorman Sean P, Gilmore Brendan F. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013;11(3):297–308. doi:10.1586/eri.13.122
2. Issakhanian L. Antimicrobial agents and urinary tract infections. Curr Pharm Des. 2019;25(12):1409–1423. doi:10.2174/1381612825999190619130216
3. Ahmadi Z, Noormohammadi Z, Ranjbar R, et al. Prevalence of tetracycline resistance genes tet (A, B, C, 39) in Klebsiella pneumoniae isolated from Tehran, Iran by multiplex PCR. Iran J Med Microbiol. 2022;16(2):141–147. doi:10.30699/ijmm.16.2.141
4. Ahmadi Z, Noormohammadi Z, Behzadi P, et al. Molecular detection of gyrA mutation in clinical strains of Klebsiella pneumoniae. Iran J Public Health. 2022;51(10):2334–2339. doi:10.18502/ijph.v51i10.10992
5. Behzadi P, García-Perdomo HA, Karpiński TM, Issakhanian L. Metallo-ß-lactamases: a review. Mol Biol Rep. 2020;47(8):6281–6294. doi:10.1007/s11033-020-05651-9
6. Shrivastava S, Shrivastava PS, Ramasamy J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J Med Soc. 2018;32(1):76–85. doi:10.4103/jms.jms_25_17
7. World Health Organization. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter Baumannii and Pseudomonas Aeruginosa in Health Care Facilities. Geneva: World Health Organization; 2017. PMID: 29630191.Bookshelf ID: NBK493061.
8. Dickstein Y, Edelman R, Dror T, et al. Carbapenem-resistant Enterobacteriaceae colonization and infection in critically ill patients: a retrospective matched cohort comparison with non-carriers. J Hosp Infect. 2016;94(1):54–59. doi:10.1016/j.jhin.2016.05.018
9. Bush K, Bradford PA. Epidemiology of β-lactamase-producing pathogens. Clin Microbiol Rev. 2020;33(2). doi:10.1128/CMR.00047-19
10. Pons MJ, Marí-Almirall M, Ymaña B, et al. Klebsiella pneumoniae spread of ST348 producing NDM-1 in a peruvian hospital. Microorganisms. 2020;8(9):1392. doi:10.3390/microorganisms8091392
11. Zhang ZX, Chen D, Xu G, et al. Molecular epidemiology and drug resistant mechanism in carbapenem-resistant Klebsiella pneumoniae isolated from pediatric patients in Shanghai, China. PLoS One. 2018;13(3):e0194000. doi:10.1371/journal.pone.0194000
12. Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–193. doi:10.1128/CMR.15.2.167-193.2002
13. Diancourt L, Passet V, Verhoef J, et al. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi:10.1128/JCM.43.8.4178-4182.2005
14. Su S, Zhang J, Zhao Y, et al. Outbreak of KPC-2 carbapenem-resistant Klebsiella pneumoniae ST76 and carbapenem-resistant K2 hypervirulent Klebsiella pneumoniae ST375 strains in Northeast China: molecular and virulent characteristics. BMC Infect Dis. 2020;20(1):472. doi:10.1186/s12879-020-05143-y
15. Zheng R, Zhang Q, Guo Y, et al. Outbreak of plasmid-mediated NDM-1-producing Klebsiella pneumoniae ST105 among neonatal patients in Yunnan, China. Ann Clin Microbiol Antimicrob. 2016;15(1):10. doi:10.1186/s12941-016-0124-6
16. Qin X, Wu S, Hao M, et al. The colonization of carbapenem-resistant Klebsiella pneumoniae: epidemiology, resistance mechanisms, and risk factors in patients admitted to intensive care units in China. J Infect Dis. 2020;221(Suppl 2):S206–S214. doi:10.1093/infdis/jiz622
17. Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18. doi:10.1186/s12941-017-0191-3
18. van Duin D, Arias CA, Komarow L, et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis. 2020;20(6):731–741. doi:10.1016/S1473-3099(19)30755-8
19. Prigitano A, Perrone PM, Esposto MC, et al. ICU environmental surfaces are a reservoir of fungi: species distribution in northern Italy. J Hosp Infect. 2022;123:74–79. doi:10.1016/j.jhin.2022.02.006
20. Durán-Manuel EM, Cruz-Cruz C, Ibáñez-Cervantes G, et al. Clonal dispersion of Acinetobacter baumannii in an intensive care unit designed to patients COVID-19. J Infect Dev Ctries. 2021;15(1):58–68. doi:10.3855/jidc.13545
21. Cao T, Liu Y, Li Y, et al. A public health concern: emergence of carbapenem-resistant Klebsiella pneumoniae in a public transportation environment. J Antimicrob Chemother. 2020;75(10):2769–2772. doi:10.1093/jac/dkaa260
22. Qiao F, Wei L, Feng Y, et al. Handwashing sink contamination and carbapenem-resistant Klebsiella infection in the intensive care unit: a prospective multicenter study. Clin Infect Dis. 2020;71(Suppl 4):S379–S385. doi:10.1093/cid/ciaa1515
23. Feng Y, Wei L, Zhu S, et al. Handwashing sinks as the source of transmission of ST16 carbapenem-resistant Klebsiella pneumoniae, an international high-risk clone, in an intensive care unit. J Hosp Infect. 2020;104(4):492–496. doi:10.1016/j.jhin.2019.10.006
24. Meng X, Yang J, Duan J, et al. Assessing molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae (CR-KP) with MLST and MALDI-TOF in central China. Sci Rep. 2019;9(1):2271. doi:10.1038/s41598-018-38295-8
25. Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. doi:10.1093/jac/dkz446
26. Ahmadi M, Ranjbar R, Behzadi P, et al. Virulence factors, antibiotic resistance patterns, and molecular types of clinical isolates of Klebsiella Pneumoniae. Expert Rev Anti Infect Ther. 2022;20(3):463–472. doi:10.1080/14787210.2022.1990040
27. Falcone M, Tiseo G, Galfo V, et al. Bloodstream infections in patients with rectal colonization by Klebsiella pneumoniae producing different type of carbapenemases: a prospective, cohort study (CHIMERA study). Clin Microbiol Infect. 2022;28(2):298.e1–298.e7. doi:10.1016/j.cmi.2021.06.031
28. Han R, Shi Q, Wu S, et al. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314. doi:10.3389/fcimb.2020.00314
29. Chen Y, Fang L, Yang Y, et al. Emergence of carbapenem-resistant Klebsiella pneumoniae harbouring blaOXA-48-like genes in China. J Med Microbiol. 2021;70(3). doi:10.1099/jmm.0.001306
30. Sun S, Gao H, Liu Y, et al. Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae. Emerg Microbes Infect. 2020;9(1):1102–1113. doi:10.1080/22221751.2020.1768805
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.