Back to Journals » The Application of Clinical Genetics » Volume 14
Evaluation of NLRP3 (rs10754558) and PTPN22 (1858C/T) (rs2476601) Functional Polymorphisms in Psoriasis Susceptibility in Egypt
Authors ALrefai A , Dawood A, Shehata W , Elhelbawy M, Elhelbawy N
Received 7 May 2021
Accepted for publication 8 July 2021
Published 26 July 2021 Volume 2021:14 Pages 331—339
DOI https://doi.org/10.2147/TACG.S319065
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Prof. Dr. Martin Maurer
Abeer ALrefai,1,2 Ashraf Dawood,1 Wafaa Shehata,3 Mohammed Elhelbawy,4 Nesreen Elhelbawy1
1Medical Biochemistry and Molecular Biology Department, Faculty of Medicine, Menoufia University, Shebin Elkom City, Egypt; 2Biochemistry Department, Faculty of Medicine, Umm Al-Qura University, Makah City, Saudi Arabia; 3Dermatology, Andrology & Sexually Transmitted Diseases (STDs) Department, Faculty of Medicine, Menoufia University, Shebin Elkom City, Egypt; 4Clinical Pathology Department, Faculty of Medicine, Menoufia University, Shebin Elkom City, Egypt
Correspondence: Nesreen Elhelbawy Tel +2 01069590533
Fax +20482181009
Email [email protected]
Background: Psoriasis is a complex autoimmune multifactorial disease induced by interaction of environmental and genetic factors. This research aimed to clarify the association of NLRP3 (rs10754558) and PTPN22 (1858C/T) (rs2476601) polymorphisms with susceptibility to psoriasis.
Methods: This case–control study involved 150 patients diagnosed with psoriasis and 100 age- and gender-matched apparently healthy individuals. NLRP3 (rs10754558) polymorphism was done by real time PCR and PTPN22 1858C/T (rs2476601) genotype was identified by tetra-primer amplification refractory mutation system-polymerase chain reaction (PCR) method.
Results: The genotypes distribution of NLRP3 (rs10754558) were significantly associated with psoriasis (p< 0.0001). Whereas for PTPN22 (1858C/T) (rs2476601), no significance was found (p=0.09). NLRP3 (rs10754558) GC genotype revealed a significant association with psoriasis (p< 0.0001), mainly among male (p=0.004) patients with mild psoriasis (p=0.001) and affected extremities (p=0.0001).
Conclusion: We can conclude that the NLRP3 (rs10754558) GC genotype may play a role in psoriasis susceptibility among male Egyptian populations with affected extremities. Future studies must evaluate its role in the prevention or the treatment of psoriasis.
Keywords: psoriasis, NLRP3 (rs10754558), PTPN22 (1858C/T) (rs2476601), real time PCR
Introduction
Psoriasis is a chronic multisystem autoimmune disease.1 It affects 1–3% worldwide populations varying among different ethnic populations and geographical regions.2 Although the etiopathogenesis of psoriasis is not fully understood, it is considered as a disease with multi-factorial etiology including different genetic (either immune-specific or skin-specific genes) and non-genetic factors, such as immunological (The imbalance among CD4+ T-cell subsets contributes to hyperkeratosis and parakeratosis) and environmental risk factors (UV radiation exposure, some medications, smoking, alcohol intake, infections, and stress). Nowadays the interaction between all these factors is the main factor in the pathogenesis of psoriasis.3 NLRP3 (NOD-like receptor family, pyrin domain-containing subfamily) is activated in skin inflammatory diseases, but its attribution in psoriasis is still questioned. The role of NLRP3 inflammasome in psoriasis has received widespread attention and recognition. Variable studies have indicated that activation of NLRP3 inflammasome may contribute to psoriasis inflammatory response.4,5 The relationship between NLPR3 and psoriasis from the perspective of genetic polymorphisms demonstrated that two single nucleotide polymorphisms (SNPs), rs3806265 and rs10754557, in NLRP3 were significantly related to Psoriasis Vulgaris (PsV).5 Protein tyrosine phosphatase non-receptor 22 (PTPN22) gene, presented on chromosomes 1p13.3 to p13.1, encodes a lymphoid-specific phosphatase (Lyp). It was found that PTPN22 interacts with and dephosphorylates NLRP3 upon proinflammatory insults, permitting robust NLRP3 activation and interleukin (IL-1β) secretion.6 It targets different signaling intermediates involved in T-cell receptor signaling and may work at many levels in the signaling cascade.7 Single nucleotide polymorphism (SNP) in PTPN22 gene 1858C/T (rs2476601) (exon 14) has been associated with the number of autoimmune diseases and considered as a risk factor due to induction of auto antibodies production.8 A limited number of studies have evaluated the association of NLPR3 genetic locus (rs10754558) with psoriasis. To explore this important issue, we investigated the association of NLRP3 (rs10754558) and PTPN22 (1858C/T) (rs2476601) polymorphisms with susceptibility to psoriasis.
Materials and Methods
Subjects
This case-control study involved 150 patients diagnosed with psoriasis in the dermatology clinic of Menoufia University Hospital and 100 age- and gender-matched apparently healthy people served as the control group. They were examined and diagnosed by dermatologists depending on the clinical findings, skin changes, and the location of the condition on the body and skin biopsy (when indicated). The demographic data and Psoriasis Area and Severity Index (PASI) score were recorded (PASI score was determined following the work by Fredriksson and Pettersson; mild psoriasis: PASI is <10, moderate psoriasis: PASI is 10–20, severe psoriasis: PASI is >20).9 All patients diagnosed with plaque psoriasis for at least 1 year were included in the study. Exclusion criteria were coexisting or family history of any other dermatological disorders (controls also), hypothyroidism, autoimmune diseases and renal/liver failure. This study was approved by the Ethical Committee, Faculty of medicine Menoufia University and performed in accordance with the Declaration of Helsinki. All participants were informed and signed consent forms before being included. NLRP3 (rs10754558) polymorphism was done by real time PCR and PTPN22 1858C/T (rs2476601) genotype was identified by tetra-primer amplification refractory mutation system-polymerase chain reaction (PCR) method.
Methods
Five milliliters of venous blood were collected in an EDTA tube for DNA extraction and genotyping of NLRP3 (rs10754558) and PTPN22 1858C/T (rs2476601).
DNA Extraction and Genotyping
Genomic DNA was extracted and purified from whole blood utilizing QIAamp DNA Mini Kit (QIAGEN, USA, 2012)10 according to the manufacturer’s protocol. Purified DNA was quantified using the Nano-Drop spectrophotometer and stored at −20 until analysis.
NLRP3 (rs10754558) G>C SNP was identified by TaqMan 5ʹ bi-allelic discrimination assay using primers and probes from Applied Biosystems (Foster City, CA, USA) and utilizing the 7500 Real-time PCR system.
The provided fluorescent labeled TaqMan probes were sequenced as: *GACAATGACAGCATCGGGTGTTGTT[C/G]TCATCACAGCGCCTCAGTTAGAGGA (VIC dye for allele C and FAM dye for allele G). To detect the selected SNPs, real-time PCR was set with a reaction volume of 25 µL using 12.5 µL of TaqMan Genotyping Master Mix, 1.25 µL of 20× SNP assay mixture of probes and primers, 6.25 µL of nuclease-free water, and 5 µL of the extracted DNA. The reaction was completed in 96-well plates and proceeded as described: 94°C for 4 minutes (pre-PCR read), then 50 cycles of 30 seconds for denaturation at 94°C, 25 seconds for primer annealing at 50°C, 40 seconds for extension at 72°C, and finally 3 minutes at 72°C for post-PCR.11
PTPN22 (rs2476601) 1858C/T genotypes were identified by tetra-primer amplification refractory mutation system. The genomic DNA was amplified by using two external primers (forward outer primer: 5′-CTCACACATCAG CTTCCCAAAGTG-3′ and reverse outer primer: 5′-CAACTTTACTGATAATGTTGCTTCAACGGA-3′) and two internal primers (forward inner primer: 5′-CAACCACAATAAATG ATTCAGGTGTACG-3′, reverse inner primer: 5′-ATCCCCCCTCCACTTCCTGGAT-3′). Product sizes were 213 and 151 bp (base pairs) for the C- and T-alleles, respectively, but the product size of the internal control was 314 bp. The PCR cycling conditions were as shown: 5 minutes at 95°C followed by 30 cycles (30 seconds at 95°C, 30 seconds at 64°C and 30 seconds at 72°C) and 10 minutes at 72°C. The all PCR products were electrophoresed on 2% agarose gels and photographed.12
Statistical Analysis
Data were statistically analyzed by SPSS version 16 (SPSS Chicago., Inc.). Mann–Whitney test was used for non-parametric variables. Chi-Squared and Fisher’s exact tests were for used qualitative variables. A p-value≤0.05 was considered significant.
Results
This is a case-control study involving 150 patients whose mean ages were 42.7±16.1 years; 105 (70%) were male and 45 (30%) were female, 30 (20%) of them have a family history of psoriasis, 24 (16%) were diabetics, nine (6%) were hypertensive, and 12 (8%) were combined diabetics and hypertensive. One hundred apparently healthy individuals were selected as a control group whose mean ages were 39.6±14.4 years; 60 (60%) were male and 40 (40%) were female. Both patients and control groups were matched for age (p=0.13) and gender (p=0.1). Thirty-nine (26%) of the patients were smokers versus 40 (40%) among the control group (p=0.02) (Table 1).
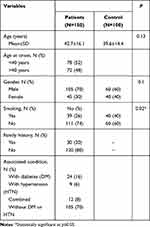 |
Table 1 Description of the Studied Groups |
A total of 81 patients (54%) had mild, 39 patients (26%) had moderate, and 30 patients (20%) presented severe psoriasis. Seventy-two (48%) of the patients suffered from early onset psoriasis and 78 (52%) of them had late onset psoriasis. Twenty-three (22%) of the psoriatic patients had a progressive course versus 12 (8%) and 105 (70%), who had stationary and regressive courses, respectively. Scalp, joint, nail, palm, and sole involvement were detected in 69 (46%), 33 (22%), 36 (24%), and 57 (38%) patients, respectively, and their median PSAI score was 8.2 (Table 2).
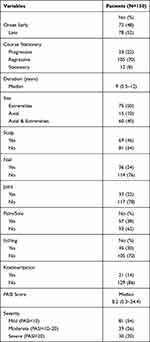 |
Table 2 Description of Psoriasis Among Patients |
Younger (p<0.0001), non-smoker (p<0.0001), with age of onset <40 years (p<0.0002) revealed significant associations with severe psoriasis. On the other hand male gender was more among patients with mild and moderate psoriasis (p=0.018). As expected, severe psoriasis was characterized by early-onset (p=0.03), progressive course (p=0.0001), itching (p=0.0003), involvement of scalp (p=0.0002), involvement of both axilla and extremities (p=0.0001), with no associated DM OR HTN (p=0.0001) (Table 3).
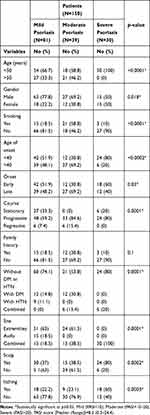 |
Table 3 Correlation Between Psoriasis Severity and Different Variables |
The genotype and allele frequencies of rs10754558 SNP of NLRP3 in the studied groups are presented in Table 4. For the control group the genotype distribution was 40 (40%) GG, 20 (20%) GC, and 40 (40%) CC; while for the patients GG was 36 (24%), GC was 81 (54%), and CC was 33 (22%). The GC genotype revealed a significant association with psoriasis (p<0.0001), while the allele showed a non-significant association with psoriasis (p=0.9). CC genotype revealed a significant decrease among patients compared to controls (p<0.0001).
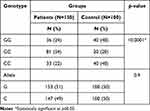 |
Table 4 Genetic Distribution of NLRP3 (rs10754558) Among Studied Groups |
The GC genotype of rs10754558 SNP was significantly associated with mild psoriasis (p=0.001) and affected extremities (p=0.0001) mainly among male patients (p=0.004) with a progressive course (p=0.03) (Table 5).
 |
Table 5 Distribution of NLRP3 (rs10754558) Genotypes Among Different Risk Groups |
On the other hand, PTPN22 1858C/T (rs2476601) genotype; CC 102 (68%), CT 47 (31.3%), and TT 1 (0.7%) in patients versus CC 80 (80%), CT 20 (20%), and TT 0 (0%) in the control group (p=0.09) revealed a non-significant association with psoriasis, while the allele frequency (p<0.05) showed critical level significance (Table 6).
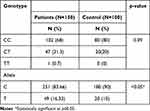 |
Table 6 Genetic Distribution of PTPN22 1858C/T (rs2476601) Genotypes Among Studied Groups |
Discussion
Psoriasis is an inflammatory immune-mediated disease with a complex genetic basis.13 Psoriasis can be correlated to multiple genes associated with linkage disequilibrium in genetically susceptible persons. Over the past four decades, by the help of genome-wide association studies (GWASs) with large cohorts, much light has been focused on the importance of different genetic factors in the pathogenesis of psoriasis.14 Previous experimental evidence has already cleared that NLRP3 inflammasome is stimulated in inflammatory skin diseases.15–17 It was proven that stimulation of NLRP3 inflammasomes leads to an inflammatory response mainly driven by increased secretion of IL1b and IL-18 that play important roles in host defense and inflammation by activating innate immune inflammatory responses.18 Furthermore, NLRP3 inflammasome plays a significant role in the maturation and activation of dendritic cells induced by MHC-II exposition on the macrophage/antigen presenting cells surface, thus dysregulation of NALP3 may impair the immune reaction.19 Genetic variants affecting the inflammasome function including NLRP3 are candidate genes that could induce susceptibility to auto-inflammatory conditions.20
Our study evaluated the relationship of NLRP3 rs10754558 and (PTPN22) 1858C/T (rs2476601) polymorphisms with susceptibility to psoriasis.
In the current study, the GC genotype of NLRP3 (rs10754558) was significantly associated with risk of psoriasis mainly among psoriatic male with mild psoriasis and affected extremities, thus underlining the importance of NLRP3 signaling in the development of psoriasis.
Conflicting results for the association of NLRP3 (rs10754558) and development of psoriasis have previously been reported.
The genotypes of NLRP3rs10754558 influenced the extent of systemic inflammation, suggesting that the SNP at the NLRP3 rs10754558 locus may modulate inflammasome activation and contribute to adverse inflammatory outcomes.21 On the other hand, Irrera et al22 confirmed the partial role of NLRP3 in the development of psoriasis. They reported that BAY 11–7082, an antagonist of NF-κB, can alleviate psoriasis-like dermatitis by inhibiting the NLRP3 inflammasome and the NF-κB pathway. Furthermore, Lee and Bae18 reported a significant association between the NLRP3 rs10754558 C/G polymorphism and both inflammatory and autoimmune diseases in Latin American individuals, but not in Europeans and Asians. However, this meta-analysis did not identify an association between it and gout and celiac disease, that could be explained by a disease-specific effect or unknown factors.18
Numerous researches have cleared the association of the gene polymorphisms of NLRP3 rs10754558 with susceptibility to diseases23 such as aspirin-induced asthma,24 abdominal aortic aneurysms, and common inflammatory disorders.25 Also, Su et al4 proved that NLRP3 is significantly upregulated in psoriatic tissues and its expression is associated with upregulation of caspase-1 leading to secretion of IL-1b and IL-18, which are involved in inflammation and immune response. The altered expression of these proteins indicates their potential role in psoriasis. Thus, NLRP3 inhibition could interrupt the pathological mechanisms underlying the triggering and the maintenance of psoriasis lesions.22
The NLRP3 rs10754558 C/G polymorphism may affect its mRNA stability and expression. The rs10754558 C allele decreases the stability of NLRP3 mRNA in comparison with the rs10754558 G allele. Moreover, the allele-specific construct containing the G allele of the rs10754558 shows a 1.3-fold higher activity than that containing the C allele.24,25 Notably, the NLRP3 rs10754558-G allele was additionally linked to high IL-18 levels, suggesting that this SNP may modulate inflammasome induction and contribute to unwanted inflammatory outcomes.21
Many autoimmune diseases can nowadays be treated with IL-1β and IL-18 antagonists or their receptors.26 However, it was found that targeting NLRP3 might present certain advantages over the use of IL-1β biologic inhibitors.27
It has been shown that the TNF-α-mediated activation of NLRP3 inflammasomes in psoriatic patients can contribute to systemic inflammation. Anti-TNF therapy normalized the inflammasome function, suggesting a mechanism for the cardiovascular risk-reducing effect.28
On the other hand, Loft et al29 reported that variants in genes involved in pattern recognition (TLRs and NOD-like receptors) pathways could not predict a response in patients treated with either anti-TNF or ustekinumab.
Several studies suggested that activation of NLRP3 is regulated by protein tyrosine phosphatase non-receptor 22 (PTPN22). Spalinger et al30 explained how loss of PTPN22 and subsequent enhanced NLRP3 phosphorylation mediate depressed NLRP3 inflammasome activation and IL1B secretion. The presence of a variant in the gene coding for PTPN22 promotes inflammasome activity. This variant is associated with increased risk to develop many chronic inflammatory disorders.
On the other hand, our study revealed a non-significant association of PTPN22 1858C/T (rs2476601) genotype with risk of psoriasis.
The PTPN22 gene encodes LYP, a functional protein tyrosine phosphatase, a regulator of the negative regulatory kinase in T-cells, inducing suppression of T-cells and, therefore, maybe the mechanism by which this association contributes to the genetic susceptibility of autoimmune disorders.31
Inconsistent with our result, Bin Huraib et al12 proved a weak association between the heterozygous CT genotype of the PTPN22 (1858C/T) gene functional variant and risk of psoriasis.Interestingly, the homozygous TT genotype was not detected at all in either patients or the controls in this study from Saudi Arabia.12
Loft et al32 ocumented a non-significant association of PTPN22 (rs2476601) as a risk factor of psoriasis arthritis (PsA). Also, Stuart et al33 could not confirm any association of this subphenotype with Psoriasis. In addition, Bowes et al34 reported a genome-wide significant association of PTPN22 (rs2476601) to PsA susceptibility, whereas no evidence for association to psoriasis. Further analysis showed an insignificant association with PTPN22 C1858T manifest in skin, the gastrointestinal tract or immune privileged sites. Revealing that the association of PTPN22 polymorphism with autoimmune diseases may depend on the localization of the affected tissue, demonstrating a role of targeted organ variation in the disease manifestations.35 Also, Chen and Chang36 showed a non-significant positive association between psoriasis and PTPN22 1858 T-allele, and the association appeared more strong among subjects with psoriatic arthritis. Recently, Al-Awadhi et al37 stated that the frequency of homozygous genotype (TT) was significantly higher in PsA patients compared to controls (p<0.0001) in Kuwaiti Arabs. However, previous studies from various geographical regions/ethnic populations on PTPN22 (+1858C/T) polymorphism and psoriasis have reported inconsistent conclusions, indicating that this association might be disease- and population-specific.
Conclusions
Our study indicated that the NLRP3 (rs10754558) GC genotype may play a role in psoriasis susceptibility among male Egyptian populations with affected extremities. Future studies must evaluate its role in the prevention or the treatment of psoriasis.
Ethical Statement
This study was conducted in accordance with the Declaration of Helsinki. Informed written consent was obtained from all the participants, approved by the Ethical Committee of Medical Research, Faculty of Medicine, Menoufia University.
Acknowledgments
We appreciate all patients and controls who contributed in this study.
Funding
There is no funding to report.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Menter A, Gottlieb A, Feldman SR, et al. Guidelines of care for the management of psoriasis and psoriatic arthritis: section 1. Overview of psoriasis and guidelines of care for the treatment of psoriasis with biologics. J Am Acad Dermatol. 2008;58:826–850. doi:10.1016/j.jaad.2008.02.039
2. Harden JL, Krueger JG, Bowcock AM. The immunogenetics of psoriasis: a comprehensive review. J Autoimmun. 2015;64:66–73. doi:10.1016/j.jaut.2015.07.008
3. Boehncke WH, Schön MP. Psoriasis. Lancet. 2015;386:983–994. doi:10.1016/S0140-6736(14)61909-7
4. Su F, Xia Y, Huang M, Zhang L, Chen L. Expression of NLPR3 in psoriasis is associated with enhancement of Interleukin-1beta and Caspase-1. Med Sci Monit. 2018;24:7909–7913. doi:10.12659/MSM.911347
5. Yu P, Hao S, Zheng H, Zhao X. Association of NLRP1 and NLRP3 polymorphisms with psoriasis vulgaris risk in the chinesehan population. Biomed Res Int. 2018;4:714–836.
6. Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood. 1999;93:2013–2024. doi:10.1182/blood.V93.6.2013.406k25_2013_2024
7. Bianco B, Verreschi IT, Oliveira KC, et al. PTPN22 polymorphism is related to autoimmune disease risk in patients with Turner syndrome. Scand J Immunol. 2010;72:256–259. doi:10.1111/j.1365-3083.2010.02438.x
8. Pradhan V, Borse V, Ghosh K. PTPN22 gene polymorphisms in autoimmune diseases with special reference to systemic lupus erythematosus disease susceptibility. J Postgrad Med. 2010;56:239–242. doi:10.4103/0022-3859.68651
9. Fredriksson T, Pettersson U. Severe psoriasis- oral therapy with a new retinoid. Dermatology. 1978;157:238–244. doi:10.1159/000250839
10. ipsogen® RT Kit Handbook. QIAGEN GmbH; 2015. Available from: http://rcostoya.com/uploads/galerias/con812/hb-1356-003-1072504-151021522-r3-ipsogen-rt-kit-ce-0315-row.pdf.
11. Afonina I, Zivarts M, Kutyavin I, Lukhtanov E, Gamper H, Meyer RB. Efficient priming of PCR with short oligonucleotides conjugated to a minor groove binder. Nucleic Acids Res. 1997;25(13):2657–2660. doi:10.1093/nar/25.13.2657
12. Bin Huraib G, Al Harthi F, Arfin M, Rizvi S, Al-Asmari A. The protein tyrosine phosphatase nonreceptor 22 (PTPN22) R620W functional polymorphism in psoriasis. Clin Med Insights Arthritis Musculoskelet Disord. 2017;11:1–6.
13. Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Ann Rev Immunol. 2014;32:227–255. doi:10.1146/annurev-immunol-032713-120225
14. Chandy S, Song J. Association of NLRP1 and NLRP3 gene polymorphism with psoriasis. Our Dermatol Online. 2020;11(3):275–283. doi:10.7241/ourd.20203.13
15. Bitto A, Altavilla D, Pizzino G, et al. Inhibition of inflammasome activation improves the impaired pattern of healing in genetically diabetic mice. Br J Pharmacol. 2014;171:2300–2307. doi:10.1111/bph.12557
16. Chiazza F, Couturier-Maillard A, Benetti E, et al. Targeting the NLRP3 inflammasome to reduce diet-induced metabolic abnormalities in mice. Mol Med. 2015;21:1025–1037. doi:10.2119/molmed.2015.00104
17. Villani A, Lemire M, Fortin G, et al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat Genet. 2009;41(1):71–76. doi:10.1038/ng285
18. Lee YH, Bae SC. Association between functional NLRP3 polymorphisms and susceptibility to autoimmune and inflammatory diseases: a meta-analysis. Lupus. 2016;25:1558–1566.
19. Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411(6837):603–606. doi:10.1038/35079114
20. Roberts RL, Van Rij AM, Phillips LV, et al. Interaction of the inflammasome genes CARD8 and NLRP3 in abdominal aortic aneurysms. Atherosclerosis. 2011;218(1):123–126. doi:10.1016/j.atherosclerosis.2011.04.043
21. Ravimohan S, Nfanyana K, Tamuhla N, Tiemessen CT, Weissma D, Bisson GP. Common variation in NLRP3Is associated with early death and elevated inflammasome biomarkers among advanced HIV/TB co-infected patients in Botswana. Open Forum Infect Dis. 2018;5:75. doi:10.1093/ofid/ofy075
22. Irrera N, Vaccaro M, Bitto A, et al. BAY 11-7082 inhibits the NF-κB and NLRP3 inflammasome pathways and protects against IMQ-induced psoriasis. Clin Sci. 2017;131:487–498. doi:10.1042/CS20160645
23. Paramel GV, Sirsjö A, Fransén K. Role of genetic alterations in the NLRP3 and CARD8 genes in health and disease. Mediators Inflamm. 2015;2015:846782.
24. Hitomi Y, Ebisawa M, Tomikawa M, et al. Associations of functional NLRP3 polymorphisms with susceptibility to food-induced anaphylaxis and aspirin-induced asthma. J Allerg Clin Immunol. 2009;124(4):779–785. doi:10.1016/j.jaci.2009.07.044
25. Verma D, Lerm M, Julinder RB, Eriksson P, Söderkvist P, Särndahl E. Gene polymorphisms in the NALP3 inflammasome are associated with interleukin-1 production and severe inflammation relation to common inflammatory diseases? Arthritis Rheum. 2008;58(3):888–894. doi:10.1002/art.23286
26. Perera AP, Kunde D, Eri R. NLRP3 inhibitors as potential therapeutic agents for treatment of inflammatory bowel disease. Curr Pharm Des. 2017;23:2321–2327. doi:10.2174/1381612823666170201162414
27. Liu L, Dong Y, Ye M, et al. The pathogenic role of NLRP3 inflammasome activation in inflammatory bowel diseases of both mice and humans. J Crohns Colitis. 2017;11:737–750. doi:10.1093/ecco-jcc/jjw219
28. Verma D, Fekri SZ, Sigurdardottir G, Eding CB, Sandin C, Enerbäck C. Enhanced inflammasome activity in patients with psoriasis promotes systemic inflammation. J Invest Dermatol. 2020;141:586–595.e5. doi:10.1016/j.jid.2020.07.012
29. Loft ND, Skov L, Iversen L, et al. Associations between functional polymorphisms and response to biological treatment in Danish patients with psoriasis. Pharmacogenomics J. 2018;18:494–500. doi:10.1038/tpj.2017.31
30. Spalinger M, Lang S, Gottier C, et al. PTPN22 regulates NLRP3-mediated IL1B secretion in an autophagy-dependent manner. Autophagy. 2017;13:1590–1601. doi:10.1080/15548627.2017.1341453
31. Vang T, Congia M, Macis MD, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37(12):1317–1319. doi:10.1038/ng1673
32. Loft N, Skov L, Rasmussen M, et al. Genetic polymorphisms associated with psoriasis and development of psoriatic arthritis in patients with psoriasis. PLoS One. 2018;13:e0192010. doi:10.1371/journal.pone.0192010
33. Stuart PE, Nair RP, Tsoi LC, et al. Genome-wide association analysis of psoriatic arthritis and cutaneous psoriasis reveals differences in their genetic architecture. Am J Hum Genet. 2015;97(6):816. doi:10.1016/j.ajhg.2015.10.019
34. Bowes J, Loehr S, Budu-Aggrey A, et al. PTPN22 is associated with susceptibility to psoriatic arthritis but not psoriasis: evidence for a further PsA-specific risk locus. Ann Rheum Dis. 2015;74(10):1882–1885. doi:10.1136/annrheumdis-2014-207187 PMID: 25923216; PubMed Central PMCID: PMCPMC4602265.
35. Zheng J, Ibrahim S, Petersen F, Yu X. Meta-analysis reveals an association of PTPN22 C1858T with autoimmune diseases, which depends on the localization of the affected tissue. Genes Immun. 2012;13:641–652. doi:10.1038/gene.2012.46
36. Chen YF, Chang JS. PTPN22 C1858T and the risk of psoriasis: a meta-analysis. Mol Biol Rep. 2012;39(8):7861–7870. doi:10.1007/s11033-012-1630-z
37. Al-Awadhi A, Haider M, Sukumaran J, Mohammed A, Hasan E, Bartella Y. Role of Protein Tyrosine Phosphatase (PTPN22) gene [C1858T] functional variant in genetic susceptibility of psoriatic arthritis in Kuwaiti Arabs. Open Rheumatol J. 2020;14:15–21. doi:10.2174/1874312902014010015
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
