Back to Journals » Hepatic Medicine: Evidence and Research » Volume 13
Evaluation of MicroRNA-122 as a Biomarker for Chronic Hepatitis C Infection and as a Predictor for Treatment Response to Direct-Acting Antivirals
Authors Elabd NS , Tayel SI , Elhamouly MS, Hassanein SA, Kamaleldeen SM, Ahmed FE, Rizk M, Gadallah AA, Ajlan SE, Sief AS
Received 21 November 2020
Accepted for publication 5 February 2021
Published 15 March 2021 Volume 2021:13 Pages 9—23
DOI https://doi.org/10.2147/HMER.S292251
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Gerry Lake-Bakaar
Naglaa S Elabd,1 Safaa I Tayel,2 Moamena S Elhamouly,1 Shaimaa A Hassanein,3 Samar M Kamaleldeen,4 Fatma E Ahmed,5 Mahmoud Rizk,6 Abdelnaser A Gadallah,7 Soma E Ajlan,8 Ahmed S Sief9
1Tropical Medicine Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 2Medical Biochemistry and Molecular Biology Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 3Diagnostic Radiology Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 4Clinical Pathology Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 5Clinical Pharmacology Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 6Internal Medicine Department, Faculty of Medicine, Banha University, Banha, Egypt; 7Internal Medicine Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 8Microbiology and Immunology Department, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 9Hepatology and Gastroenterology Department, Shebin Elkom Teaching Hospital, Menoufia, Egypt
Correspondence: Safaa I Tayel
Medical Biochemistry and Molecular Biology Department, Faculty of Medicine, Menoufia University, Shebin El-Kom, Menoufia, 32511, Egypt
Tel +20 1003383097
Email [email protected]
Background: Treatment response to antiviral drugs is a challenging issue in patients with chronic hepatitis C virus (HCV) infection. Although microRNA-122 represents the majority of the microRNA content in hepatic tissues, few studies have evaluated its role in the treatment response, so we aimed to study its role in chronic HCV patients and in predicting the treatment response to direct-acting antivirals (DAAs).
Methods: The study included 125 chronic HCV patients (89 naïve and 36 with a prior failed peginterferon/ribavirin response) and 50 apparently healthy subjects. Complete blood count, liver function, α-fetoprotein, lipid profiles, serum creatinine, abdominal ultrasound, and FibroScan® were assessed. Viral markers, HCV antibodies, and hepatitis B surface antigen were measured by enzyme-linked fluorescent immunoassay, with quantitative estimation of HCV RNA and microRNA-122 levels by real-time PCR.
Results: The microRNA-122 level in HCV patients (those with a sustained virologic response 12 weeks after finishing therapy [SVR12] and non-responders) was significantly increased compared with controls and expressed more in non-responders versus SVR12 (p=0.042). ROC curve analysis of microRNA-122 for differentiating HCV patients from healthy controls revealed that a cut-off point of > 1.45 had a sensitivity of 67.20%, specificity of 94.0%, AUC=0.861, and p< 0.001; and for predicting response to treatment a cut-off point ≤ 5.66 could significantly (p=0.022) predict the occurrence of SVR, with a sensitivity of 60.34%, specificity of 66.67%, and AUC=0.729. Logistic regression analysis showed significant values for microRNA-122 in multivariate and univariate analysis for the prediction of response to DAAs.
Conclusion: The results demonstrated the possible function of microRNA-122 as an indicative tool for distinguishing chronic HCV patients from controls and in the assessment of the therapeutic reaction to DAAs.
Keywords: CHC, FibroScan, microRNA-122, DAAs
Introduction
Globally, viral hepatitis has been assessed to be the seventh most common cause of mortality.1 Nearly half of this mortality is identified with hepatitis C virus (HCV), the main reason for liver cirrhosis and hepatocellular carcinoma.2 HCV is the most prevalent etiology of chronic liver diseases in Egypt. The ongoing advances in oral direct-acting antivirals (DAAs) provide opportunities to limit HCV illness and its forward transmission.3 Although the use of DAAs is effective, not all patients effectively clear the virus.4
HCV belongs to the family Flaviviridae, and is a small, enveloped, positive-sense, single-stranded RNA virus. The viral genome is 9.6 kb long, with flanking 5′- and 3′-untranslated regions (UTRs), and contains a single open reading frame (ORF) that encodes a 3000-amino-acid polypeptide, which is cut by host and viral proteases into basic and non-basic proteins.5
MicroRNAs are small, non-coding RNA molecules (18–24 nucleotides). They participate in organizing cell proliferation, differentiation, and apoptosis.6,7 This role is related to the capacity of microRNAs to attach to certain destinations of target mRNAs at the 3′-UTR that, in turn, may produce translational constraints and/or mRNA degradation.8 Furthermore, the host microRNAs can likewise attach to the mRNAs of attacking microbes such as viruses.9 There are practical relationships between cellular microRNAs and viruses. MicroRNAs probably have basic functions in viral pathogenesis as well as in regulating viral replication, either advancing or restricting it.10
A model in this setting is the integral base pairing of microRNA-122 (the liver-specific human microRNA) to HCV genomic RNA at the 5′-UTR, which is reported to encourage viral replication inside liver cells and to stimulate viral protein translation, in addition to protecting the uncapped HCV RNA from degradation.11 Consequently, microRNA-122 has attracted considerable interest regarding its potential as a therapeutic target.
In this study, we aimed to appraise the role of microRNA-122 as a marker for chronic hepatitis C (CHC) genotype 4 infection and as an indicator of the response to DAA therapy, as well as its considerable correlations with clinical and radiological parameters.
Subjects
This prospective study was conducted in the Tropical Medicine Department, in collaboration with the Medical Biochemistry and Molecular Biology, Clinical Pathology, Medical Microbiology and Immunology, Diagnostic Radiology, Clinical Pharmacology, and Internal Medicine Departments, at the Faculty of Medicine, Menoufia University, Egypt. Participants were selected from patients attending the Tropical Medicine Department between June 2017 and October 2018. In total, 175 participants were included in the study: 125 CHC cases (89 naïve to treatment and 36 with a prior failed response to peginterferon and ribavirin [pegIFN/RBV; treatment-experienced]), and 50 HCV-negative, seemingly healthy controls matched for age and gender. The sample size was calculated by a continuous response variable from independent control and experimental subjects, with 0.5 controls per experimental subject.12 Subjects were analyzed by identifying HCV antibodies, then confirmed by real-time PCR. HCV patients received treatment in the form of sofosbuvir 400 mg/day and daclatasvir 60 mg/day with or without ribavirin (weight-based ribavirin 1200 or 1000 mg/day if ≥75 or <75 kg body weight, respectively) for 12 weeks, according to the accepted treatment recommendation from the European Association for the Study of the Liver (EASL)13 and national guidelines.
Patients with these criteria were incorporated in this research: age more than 18 years, positive HCV antibodies that were confirmed by PCR for HCV-RNA and HCV genotype 4, treatment-naïve or prior failed responders to pegIFN/RBV (treatment-experienced), and patients with Child–Pugh score ≤8. Patients with renal illness, hepatocellular carcinoma, or any extrahepatic malignancy, hepatitis B virus (HBV) or HIV coinfection, and pregnant females, were excluded from this investigation.
For all participants, history taking, clinical evaluation, and laboratory investigations were conducted. Imaging evaluation was performed for all participants, including abdominal ultrasound and transient elastography (FibroScan®). Aspartate aminotransferase (AST)-to-platelet ratio index (APRI) and Fibrosis-4 (FIB-4) scores were estimated for all participants. The APRI was estimated by the following formula: (AST/upper limit of normal)/platelet count (platelets ×109/L) × 100,14 and the FIB-4 score was estimated by the formula: Age (years) × AST (IU/L)/(platelet count (×109/L) ×√ALT (IU/L)).15
During antiviral treatment, follow-up was conducted by clinical assessment and re-evaluation of laboratory tests (complete blood count [CBC], liver and kidney function tests) at weeks 4, 8, and 12 of treatment. At the end of treatment, RT-PCR was used to determine the response to treatment, together with transient elastography. Then all of the above-mentioned clinical, laboratory, and imaging investigations were performed for all patients 12 weeks after stopping treatment, including RT-PCR to determine the sustained virologic response 12 weeks after finishing therapy (SVR12) and microRNA-122.
According to their response to HCV treatment, patients were divided into two groups. Group I (GI) comprised 116 patients who achieved SVR12, defined as undetectable HCV-RNA 12 weeks after finishing therapy; it included 83 (71.55%) treatment-naïve patients and 33 (28.45%) with a prior failed response to pegIFN/RBV (treatment-experienced). Group II (GII) comprised nine patients not achieving SVR12 (non-responders); it included six (66.67%) treatment-naïve and three (33.33%) treatment-experienced patients. In addition, Group III (GIII) comprised 50 HCV antibody-negative apparently healthy participants of matched age and gender as a control group.
This investigation was conducted in accordance with the Declaration of Helsinki. Notified assent guaranteed by the ethical board of the Faculty of Medicine, Menoufia University, was acquired from all contributors, and all participants provided informed consent.
Methods
Blood Sampling
Venous blood samples (10 mL) were drawn from all subjects. Serum was separated from 5 mL of blood in a plain tube, left for 30 min to coagulate, and then centrifuged at 4000 rpm for 10 min; further colorimetric estimation of creatinine was performed utilizing a Diamond Diagnostics Kit (Germany), serum ALT, AST, and alkaline phosphatase (ALP) by the kinetic UV-optimized method IFCC LTEC Kit (UK), serum albumin by enhanced specificity of bromocresol green using a Diamond Diagnostics Kit (Germany), and total and direct bilirubin using a Diamond Diagnostics Kit (Germany). The colorimetric determination of serum total cholesterol (TC) and triglyceride (TG) levels was carried out using the Spinreact kit (Spain), and serum high-density lipoprotein cholesterol (HDLc) level was assayed using a human kit (Germany). Low-density lipoprotein cholesterol (LDLc) values were estimated by the equation (LDLc) = TC – (TG/5 + HDLc), as TG levels did not exceed 400 mg/dL. Serological markers of HCV antibodies and hepatitis B virus surface antigen (HBsAg) were detected by enzyme-linked fluorescent immunoassay (ELIFA) utilizing MINI VIDAS systems (bioMerieux, Marcy l’Etoile, France); positive tests were confirmed by real-time PCR with a threshold of detection level of 15 IU/mL and HCV genotyping. Fresh blood (2 mL) was collected in tubes containing ethylenediamine tetra-acetic acid (EDTA) and was used for a complete blood count (CBC) by Sysmex XN-1000 (Japan; BM Egypt Company). In addition, 1 mL of fresh blood was taken in a citrate tube for measurement of prothrombin time (PT) and international normalized ratio (INR) (LlQUIPLASTIN diagnostic kit, Biomed, Germany), and 2 mL of fresh blood was collected in EDTA-containing tubes and used for microRNA extraction and quantification by real-time PCR.
Estimation of MicroRNA-122 Levels
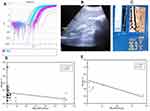
MicroRNA was extracted from whole blood using a Direct-zol™ RNA MiniPrep kit (Zymo Research) and stored at –80°C. With the aid of the TaqMan® MicroRNA Reverse Transcription Kits (Applied Biosystems), reverse transcription (RT) was performed on the extracted microRNA to synthesize single-stranded cDNA. The looped-primer RT-PCR supplied in the TaqMan MicroRNA assays can precisely identify mature microRNA. The total volume of 15 μL RT contains: 3 μL of RT primer, 5 μL miRNA, and 7 μL RT master mix, containing (10× Reverse Transcription Buffer, 100 mM dNTPs [with dTTP], RNase inhibitor 20 U/μL, MultiScribe™ Reverse Transcriptase 50 U/μL, and nuclease-free water). The 2720 Thermal Cycler (Applied Biosystems, Singapore) was programmed at 16°C for 30 min, 42°C for 30 min, 85°C for 5 min, and 4°C for ∞ for one cycle, and the RT product was preserved at –20°C. The microRNA-122 primers and TaqMan probes were designed by Applied Biosystems (Foster City, CA, USA) Life Technologies. The total 20 μL of PCR master mix comprised 10 μL 2× TaqMan Universal Master mix, 7 μL RNase-free water, 1.0 μL 20× TaqMan MicroRNA Primer Assay mix, and 2 μL of RT product, and RNU48 was utilized as an internal control. The 7500 Thermal Cycler (Applied Biosystems, Foster City, CA, USA) was set up as follows: first PCR initial activation step at 50°C for 2 min hold and 95°C for 10 min hold, then two-step cycling of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min, for 40 cycles. The comparative ∆∆Ct method was applied for data analysis to calculate the relative quantification (RQ), where microRNA-122 in the samples was normalized to a housekeeping gene (RNU48) and relative to a control (Figure 1A).
HCV Genotyping Using the Nested PCR/Ohno Method
A Qiagen Viral RNA kit (Hilden, Germany) was applied to separate RNA from serum. Ohno’s method was used for genotyping; this involves nested PCR amplification of the virus core gene using genotype-specific primers, as described previously.16
Imaging
Ultrasound of the Abdomen
All patients underwent abdominal ultrasonography using a 3.5–5 MHz probe. The liver was scanned in the axial and longitudinal views while the patient was supine (Figure 1B) and the patency and size of the portal vein were assessed in left lateral decubitus. Patients were also assessed in the supine and right lateral decubitus positions for spleen size, measured as the bipolar diameter.
FibroScan Assessment
All patients were examined in the dorsal decubitus with the right arm in maximal abduction, using a 5 MHz ultrasound transducer probe mounted on the axis of a vibrator of the FibroScan machine (Echosens, Paris, France). The transducer tip was covered with a drop of gel and measurements were taken on the right lobe of the liver by positioning the tip of the transducer perpendicularly in the intercostal space. A cylinder of hepatic tissue measuring about 1 cm in diameter and 2–4 cm in length was then chosen, avoiding any large vascular structures within. This was performed under ultrasound time–motion imaging (Figure 1C).
Statistical Analysis
Data were fed into a computer and analyzed using the IBM SPSS software package version 20.0 (IBM Corp, Armonk, NY, USA). To verify the normality of the distribution of variables, the Kolmogorov–Smirnov test was used. Spearman coefficients were used to assess correlations between quantitative variables. Receiver operating characteristics (ROC) curves were used to determine the diagnostic performance of microRNA-122. For comparison between different periods, the paired t-test was applied, while the Wilcoxon signed ranks test was applied for comparison between different periods for abnormally distributed quantitative variables. Regression analysis was used to detect the most independent factors and those affecting mortality. The significance of the data was assessed at the 5% level.
Results
This study was conducted on 125 CHC patients who had received DAA treatment. They were 73 males (58.4%) and 52 females (41.6%), with a mean±SD age of 47.3±12.2 years. According to the therapeutic response, patients were classified into two groups. GI, patients who achieved SVR12, included 116 patients (92.8%), 69 males (59.5%) and 47 females (40.5%), with a mean age of 47.3±12.2 years; and GII, comprising patients who did not achieve SVR12, included nine cases (7.2%), four males (44.4%) and five females (55.6%), and their mean age was 47.4±12.9 years. In addition, GIII included 50 apparently normal individuals.
We investigated the studied laboratory parameters twice, before (pre) and after (post) the treatment regimen. Data analysis was performed among the studied groups and explored the following results. Age and gender were matched among groups (p>0.05). Pre-treatment, platelets were distinctly lower in patient groups than in controls (p<0.001), with no critical distinction between GI and GII (p=0.771), while hemoglobin (Hb) concentration and white blood cells (WBCs) did not differ among groups (p=0.135 and p=0.594, respectively). With respect to liver function, total and direct bilirubin, ALT, AST (p<0.001), and INR (p=0.007) were markedly higher, while albumin (p=0.011) was lower in GI and GII than in GIII; moreover, comparison between GI and GII revealed no significant difference. We noticed lower values of TC, TG, LDLc, and HDLc in patients compared to controls. MicroRNA-122 levels exhibited marked variations (p<0.001) among groups, with distinctly higher levels in non-responders than in SVR12 (p=0.042) and controls (p<0.001) and higher levels in SVR12 than in controls (p<0.001). Regarding the assessment of fibrosis in the study participants, FibroScan (p<0.001), FIB-4 (p<0.001), and APRI score (p<0.001) were higher in the patient groups (GI and GII) than in normal participants, but did not differ between GI and GII (Table 1).
 |
Table 1 Comparison Between the Three Studied Groups According to Demographic and Laboratory Data and Fibrosis Assessment Before Treatment |
Table 2 shows that portal vein diameter, spleen diameter, and liver stiffness assessed by FibroScan did not vary between patient groups before and after treatment.
 |
Table 2 Imaging Findings in Patient Groups (Pre and Post Treatment) |
Comparisons of the measured parameters pre- and post-treatment in GI showed that platelets (p<0.001), TC (p<0.001), LDLc (p<0.018), and HDLc (p<0.001) were significantly increased in patients after reaching SVR12. Moreover, ALT, AST, PT, INR (p<0.001), microRNA-122 (p<0.001), FibroScan (p<0.001), FIB-4 (p<0.011), and APRI score (p<0.001) were diminished in SVR12 patients after treatment. Hb (p=0.089), WBC (p=0.080), total bilirubin (p=0.137), direct bilirubin (p=0.615), albumin (p=0.092), and TG (p=0.059) were not significantly changed pre- and post-treatment. Furthermore, comparison of the studied parameters pre- and post-treatment in GII revealed that in non-responders there were some small changes in laboratory and imaging findings that were not statistically significant (Table 3).
 |  |  |
Table 3 Comparison Between Pre and Post Treatment Results in Patient Groups According to Laboratory and Imaging Data |
Table 4 presents the results of a correlation analysis between microRNA-122 and the studied parameters prior to treatment in GI and GII, where we observed meaningful negative correlations between microRNA-122 and platelets using the Spearman coefficient (r=−0.265, p=0.004 and r=−0.745, p=0.021, respectively) (Figure 1D and E), and significant positive correlations with ALT (r=0.403, p<0.001 and r=0.750, p=0.02, respectively) (Figure 2A and B) and INR (r=0.215, p=0.021 and r=0.842, p=0.004, respectively); however, no associations were observed with other laboratory or imaging findings.
 |
Table 4 Correlations Between MicroRNA-122 (Pretreatment) and Laboratory Findings (Pretreatment) in Patient Groups (GI and GII) |
Table 5 shows the results of multivariate and univariate regression analysis, which demonstrated that microRNA-122 could be a significant independent biomarker for forecasting the reaction to DAA-based therapy in CHC patients in both multivariate analysis (unstandardized coefficient B=−1.026, p=0.024) and univariate analysis (B=−0.954, p=0.031).
 |
Table 5 Multivariate Analysis for the Parameters Affecting Response to Treatment |
ROC curve analysis of microRNA-122 for distinguishing patients with SVR12 from non-responders revealed that a cut-off point of ≤5.66 significantly (p=0.022) and early (before starting treatment) could predict the incidence of SVR in CHC patients receiving DAA treatment, with a sensitivity of 60.34%, specificity 66.67%, positive predictive value (PPV) 95.90%, negative predictive value (NPV) 11.50%, and area under the curve (AUC)=0.729 (Table 6, Figure 2C).
 |
Table 6 Agreement (Sensitivity, Specificity) for MicroRNA-122 to Predict SVR12 (Cases vs Non-Responders) and to Predict Chronic HCV Infection (HCV Cases vs Control) |
Moreover, ROC curve analysis of microRNA-122 for differentiating HCV-positive patients from healthy participants revealed that a cut-off point of >1.45 had a sensitivity of 67.20%, specificity 94.0%, PPV 96.60%, NPV 53.40%, AUC=0.861, and p<0.001, providing additional evidence for the position of microRNA-122 as a diagnostic biomarker for distinguishing CHC patients from healthy individuals (Table 6, Figure 2D).
Discussion
Worldwide, HCV is a major disease burden; in 2015, the prevalence was 1.0%.17 The control of chronic HCV infection has been improved with the introduction of DAAs.
In our study, 125 chronic HCV patients received DAA therapy for 12 weeks, of whom 92.8% achieved SVR12. These results are near to those obtained by Ahmed et al,18 who reported that SVR12 was achieved by 96% of a cohort of Egyptian patients with chronic HCV genotype 4.
In the current study, we observed that achieving SVR12 was associated with elevated platelet count and lower ALT and AST levels; moreover, we found an important reduction in liver stiffness on the FibroScan results. Stasi et al also detected a reduction in liver stiffness and hence the FibroScan results after DAA therapy.19 Similarly, our results showed a lowering of FIB-4 and APRI scores, which agreed with previous reports.20,21
In the normal liver parenchyma, microRNA-122 is considered the most plentiful microRNA, accounting for >70% of the total microRNA in hepatocytes and a dominant actor in liver development, differentiation, homeostasis, and metabolic functions.22 The attachment of microRNA-122 to the 5′-UTR of HCV genomic RNA is vital for HCV replication; it enhances viral protein translation and safeguards the uncapped viral RNA from degradation.23,24
To our knowledge, this study is the first to investigate the level of microRNA-122 in CHC genotype 4 patients and its relationship with the reaction to DAA therapy. Patients were classified following treatment into SVR12 and non-responder groups. We noticed a marked elevation in microRNA-122 in both SVR12 and non-responder groups compared with healthy individuals in the pretreatment stage, while in the post-treatment stage microRNA-122 levels were markedly decreased in the SVR12 group, reaching values close to those in healthy individuals. The microRNA-122 levels in the non-responder group remained high and did not differ significantly from values in the pretreatment stage. ROC curve analysis showed that microRNA-122 at a cut-off point of ≤5.66 could precisely distinguish SVR12 from non-responder patients, with a sensitivity of 60.34% and specificity of 66.67%, suggesting the potential role of microRNA-122 in evaluating and predicting the response to therapy in CHC patients.
Previous reports support our findings. Waring et al25 measured microRNA-122 expression levels in HCV genotype 1–3 patients who received IFN-free regimens, and found that microRNA-122 levels were diminished after therapy in the SVR group and that microRNA-122 showed steady alterations to the DAA regimen in genotype 1–3, that did not change with RBV intake. Amr et al,26 in a study using a sofosbuvir and pegIFN/RBV regimen in an Egyptian population, found that microRNA-122 was higher in patients than in controls pretreatment, and markedly down-regulated post-treatment.
We observed that microRNA-122 levels were increased in the non-responder group compared to SVR12 patients. However, it has previously been reported that pretreatment circulating microRNA-122 levels were up-regulated more in the SVR than in the NR group in HCV genotype 1 and 2- as well as genotype 4-infected Taiwanese and Egyptian patients, respectively, and it was proved by regression analysis that microRNA-122 was independently related to the end of regimen response in the SVR.27,28 We suggest that this inconsistency may be due to the different treatment strategies used; in both of those studies treatment was based on interferon, whereas in our study we used DAA-based therapy.
The present results showed that a cut-off point of >1.45 can distinguish chronic HCV-positive cases from healthy individuals with a sensitivity of 67.20%, specificity of 94.0%, and AUC=0.861. These results prove the potential of microRNA-122 in the diagnosis of CHC.
A meta-analysis carried by Zhou et al29 demonstrated that circulating microRNA-122 achieved a sensitivity of 0.92, specificity of 0.84, and AUC=0.93, denoting average test performance for the diagnosis of chronic viral hepatitis related to HBV and HCV, and concluded that serum microRNA-122 has a comparatively high diagnostic value, especially in CHC patients. Furthermore, Butt et al30 found that circulating microRNA-122 had a durable ability to distinguish CHC patients from healthy subjects at a cut-off value of 10.32, with 87.0% sensitivity and 96.7% specificity. Another notable finding that they reported was that microRNA-122 was elevated in CHC genotype 3 patients with either normal or elevated ALT levels compared to controls, which could be indicative of liver damage induced by chronic HCV infection rather than ALT, which is supposed to rise with disease severity. Moreover, it was previously shown that microRNA-122 in the blood increases earlier than ALT upon toxic liver injury.31,32
The results of a correlation analysis of serum microRNA-122 and ALT showed a clear significant relationship of microRNA-122 with ALT in pretreatment SVR and non-responder groups, indicating that serum microRNA-122 could reflect HCV-induced liver inflammation. These results agree with Bihrer et al,33 who reported a significant relationship of microRNA-122 with increased ALT and inflammatory scores, whereas Wang et al22 did not report any association of serum microRNA-122 with the clinical data in CHC genotype 1.
We detected no significant relationship between microRNA-122 and liver stiffness (FibroScan), FIB-4, or APRI scores. Jopling et al11 found that plasma microRNA-122 had a strong correlation with the degree of fibrosis by liver biopsy (higher level of microRNA-122 in F3 and F4 than in F2) in HCV genotype-4 patients. In addition, they documented no correlation between microRNA-122 and APRI score. Wang et al found a reverse association between microRNA-122 and fibrosis scores.22
The current study detected in multivariate and univariate regression analysis that microRNA-122 could be a significant independent biomarker for the initial forecast of treatment response in CHC disease. Waring et al25 confirmed this in a logistic regression model, from which they found that in HCV genotypes 1–3, standardized microRNA-122 baseline values could predict SVR status.
Remarkably, silencing of microRNA-122 that is markedly expressed following HCV infection and forecasts the reaction an IFN regimen11,34 leads to suppression of the expression of suppressors of cytokine signaling (SOCS3) by promoter methylation, and eventually improves the activation of signals of transduction and activators of transcription (STAT3)35 to increase the efficacy of the IFN regimen.
Miravirsen (SPC3649) is an antisense oligonucleotide, which, in a phase 2a study, demonstrated a distinguishable extensively dose-dependent reduction in HCV RNA values in CHC genotype 1.36
Conclusion
The results demonstrate the remarkable role of microRNA-122 as a diagnostic biomarker for distinguishing patients with chronic HCV from controls and in the prediction of the treatment response to DAAs. These findings may encourage the future use of antisense oligonucleotide targeting microRNA-122 in the control of HCV infection.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Stanaway JD, Flaxman AD, Naghavi M, et al. The global burden of viral hepatitis from 1990 to 2013: findings from the global burden of disease study 2013. Lancet. 2016;388(10049):1081–1088. doi:10.1016/S0140-6736(16)30579-7
2. Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57(4):1333–1342. doi:10.1002/hep.26141
3. Ayoub HH, Abu-Raddad LJ. Impact of treatment on hepatitis C virus transmission and incidence in Egypt: a case for treatment as prevention. J Viral Hepat. 2016;31: 12671.
4. Parigi TL, Torres MCP, Aghemo A. Upcoming direct acting antivirals for hepatitis C patients with a prior treatment failure. Clin Mol Hepatol. 2019;25(4):360–365. doi:10.3350/cmh.2019.0022
5. Bonkovsky HL, Mehta S. Hepatitis C: a review and update. J Am Acad Dermatol. 2001;44(2):159–182. doi:10.1067/mjd.2001.109311
6. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23(1):175–205. doi:10.1146/annurev.cellbio.23.090506.123406
7. Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15(5):563–568. doi:10.1016/j.gde.2005.08.005
8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
9. Roberts AP, Lewis AP, Jopling CL. The role of microRNAs in viral infection. Prog Mol Biol Transl Sci. 2011;102:101–139. doi:10.1016/B978-0-12-415795-8.00002-7
10. Ye X, Zhang HM, Qiu Y, et al. Coxsackievirus-induced miR-21 disrupts cardiomyocyte interactions via the downregulation of intercalated disk components. PLoS Pathog. 2014;10(4):e1004070. PMID: 24722419. doi:10.1371/journal.ppat.1004070
11. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309(5740):1577–1581. doi:10.1126/science.1113329
12. Dupont WD, Plummer WD. Power and sample size calculations: a review and computer program. Control Clin Trials. 1990;11(2):116–128. doi:10.1016/0197-2456(90)90005-M
13. EASL. Recommendations on treatment of hepatitis C. J Hepatol. 2017;66(1):153–194. doi:10.1016/j.jhep.2016.09.001
14. Wai CT, Greenson JK, Fontana RJ, et al. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;38(2):518–526. doi:10.1053/jhep.2003.50346
15. Sterling RK, Lissen E, Clumeck N, et al. APRICOT clinical investigators. development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43(6):1317–1325. doi:10.1002/hep.21178
16. Ohno O, Mizokami M, Wu R, et al. New hepatitis c virus (hcv) genotyping system that allows for identification of hcv genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35(1):201–207. [PMID: 8968908]. doi:10.1128/JCM.35.1.201-207.1997
17. World Health Organization (WHO). Hepatitis C fact sheet; July 2016. [Google Scholar]
18. Ahmed OS, Safwat E, Khalifa MO, et al. Sofosbuvir plus daclatasvir in treatment of chronic hepatitis C genotype 4 infection in a cohort of Egyptian patients: an experiment the size of Egyptian village. Int J Hepatol. 2018. doi:10.1155/2018/9616234
19. Stasi C, Piluso A, Arena U, et al. Evaluation of the prognostic value of liver stiffness in patients with hepatitis C virus treated with triple or dual antiviral therapy: a prospective pilot study. World J Gastroenterol. 2015;21(10):3013–3019. doi:10.3748/wjg.v21.i10.3013
20. Tada T, Kumada T, Toyoda H, et al. Improvement of liver stiffness in patients with hepatitis C virus infection who received direct-acting antiviral therapy and achieved sustained virological response. J Gastroenterol Hepatol. 2017;32(12):1982–1988. doi:10.1111/jgh.13788
21. Knop V, Hoppe D, Welzel T, et al. Regression of fibrosis and portal hypertension in HCV-associated cirrhosis and sustained virologic response after interferon-free antiviral therapy. J Viral Hepat. 2016;23(12):994–1002. doi:10.1111/jvh.12578
22. Wang JH, Jiang D, Rao HY, Zhao JM, Wang Y, Wei L. Absolute quantification of serum microRNA-122 and its correlation with liver inflammation grade and serum alanine aminotransferase in chronic hepatitis C patients. Int J Infect Dis. 2015;30:52–56. doi:10.1016/j.ijid.2014.09.020
23. Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4(1):77–85. doi:10.1016/j.chom.2008.05.013
24. Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84(13):6615–6625. doi:10.1128/JVI.00417-10
25. Waring JF, Dumas EO, Abel S, et al. Serum miR-122 may serve as a bioma rker for response to direct acting antivirals: effect of paritaprevir/R with dasabuvir or ombitasvir on miR-122 in HCV-infected subjects. J Viral Hepat. 2016;23(2):96–104. doi:10.1111/jvh.12470
26. Amr KS, Ezzat WM, Elbatae H, Atia H, Reda E, Elhosary Y. Assessment of serum level of miRNAs before and after treatment with sofosbuvir in Egyptian patients with chronic HCV infection. Meta Gene. 2018;18:93–99. doi:10.1016/j.mgene.2018.08.005
27. Su TH, Liu CH, Liu CJ, et al. Serum microRNA-122 level correlates with virologic responses to pegylated interferon therapy in chronic hepatitis C. Proc Natl Acad Sci U S A. 2013;110(19):7844–7849. doi:10.1073/pnas.1306138110
28. Motawi TM, Rizk SM, Shaker OG, Mokhtar OZ. MicroRNAs as predictor markers for response to interferon treatment of chronic hepatitis C genotype-4 in Egyptian patients. PLoS One. 2015;10(3):e0121524. doi:10.1371/journal.pone.0121524
29. Zhou X, Fang S, Wang M, et al. Diagnostic value of circulating miRNA-122 for hepatitis B virus and/or hepatitis C virus-associated chronic viral hepatitis. Biosci Rep. 2019;39(9):20190900. doi:10.1042/BSR20190900
30. Butt AM, Raja AJ, Siddique S, et al. Parallel expression profiling of hepatic and serum microRNA-122 associated with clinical features and treatment responses in chronic hepatitis C patients. Sci Rep. 2016;6(1):21510. doi:10.1038/srep21510
31. Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106(11):4402–4407. doi:10.1073/pnas.0813371106
32. Zhang Y, Jia Y, Zheng R, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56(12):1830–1838. doi:10.1373/clinchem.2010.147850
33. Bihrer V, Friedrich-Rust M, Kronenberge B, et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 2011;106(9):1663–1669. doi:10.1038/ajg.2011.161
34. Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR–122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15(1):31–33. doi:10.1038/nm.1902
35. Yoshikawa T, Takata A, Otsuka M, et al. Silencing of microRNA-122 enhances interferon-α signaling in the liver through regulating SOCS3 promoter methylation. Sci Rep. 2012;2(1):637. doi:10.1038/srep00637
36. Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi:10.1056/NEJMoa1209026
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.