Back to Journals » Cancer Management and Research » Volume 12
Evaluation of Concordance Between Deficient Mismatch Repair and Microsatellite Instability Testing and Their Association with Clinicopathological Features in Colorectal Cancer
Authors Bai H, Wang R, Cheng W, Shen Y, Li H, Xia W, Ding Z, Zhang Y
Received 2 February 2020
Accepted for publication 8 April 2020
Published 24 April 2020 Volume 2020:12 Pages 2863—2873
DOI https://doi.org/10.2147/CMAR.S248069
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Huili Bai,1,* Rong Wang,1,2,* Wei Cheng,1 Yifan Shen,1 Haijun Li,2 Wei Xia,1 Zhenglin Ding,3,* Yuhong Zhang1,*
1The Center for Clinical Molecular Medical Detection, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 2Department of Laboratory Medicine, Guangyuan Central Hospital, Guangyuan, Sichuan, People’s Republic of China; 3Department of Laboratory Medicine, The People’s Hospital of Nanchuan, Chongqing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yuhong Zhang; Zhenglin Ding Email [email protected]; [email protected]
Background: Microsatellite instability (MSI) is one of the most important molecular characteristics of colorectal cancer (CRC), which mainly results from defective DNA mismatch repair (MMR). This study was performed to investigate the concordance between deficient MMR and MSI testing, and to evaluate the association of these two results with clinicopathological characteristics in Chinese CRC patients.
Methods: A total of 738 CRC patients were included. Tumor tissues and paired peripheral blood specimens were obtained. Screening for MMR was investigated using immunohistochemical (IHC) technique, and multiple polymerase chain reaction-capillary electrophoresis (PCR-CE) method was performed to detect the MSI status. All clinicopathological data, immunohistochemistry and microsatellite instability analyses were then statistically analyzed.
Results: Of the 738 (17.75%) CRC patients, 131 expressed as deficient mismatch repair (dMMR) status, and postmeiotic segregation increased 2 (PMS2) deficiency was the most frequent deficiency among these four MMR proteins. MSI-high (MSI-H) status occurred in 74 of the 738 (10.03%) CRC patients, 55 of whom showed instability at all six mononucleotides repeat markers. dMMR was significantly associated with MSI-H and moderate concordance was observed between IHC and PCR-CE in evaluating deficient MMR/MSI through Kappa test. Statistically, dMMR was significantly associated with younger age, right-sided colon and poor differentiation. MSI-H was associated with younger age, right-sided colon, poor differentiation, mucinous type and tumor, node, metastasis (TNM) stage II.
Conclusion: A moderate concordance between deficient MMR and MSI testing indicates that both IHC and PCR-CE methods should be routinely tested to provide reliable data for clinical treatment decisions.
Keywords: deficient mismatch repair, microsatellite instability, clinicopathological features, colorectal cancer
Introduction
MSI is characterized by abnormal insertions/deletions in short tandem repeats, which largely results from dysfunction of MMR genes.1 In particular, the immunological checkpoint inhibitors anti-programmed cell death protein 1 (anti-PD-1) Pembrolizumab and Nivolumab were recommended by National Comprehensive Cancer Network (NCCN) clinical practice guidelines for the first time in the treatment of metastatic colorectal cancer (mCRC) with dMMR/MSI-H molecular phenotype in 2017.2 Anti-PD-1 provided durable responses and sustained disease control in dMMR/MSI-H mCRC patients. Furthermore, dMMR is often applied to diagnose Lynch syndrome (LS), a hereditary non-polyposis colorectal cancer due to germline mutations in MMR system(mutL homologue 1 (MLH1), postmeiotic segregation increased 2 (PMS2), mutS homologue 2 (MSH2) and 6 (MSH6) proteins) but silencing with BRAF mutation.3 Additionally, MMR status has been demonstrated to provide valuable prognostic analysis.4,5 dMMR/MSI-H have been mainly observed within CRC and endometrial cancer.6
CRC is the third most commonly diagnosed cancer and takes the fourth place as a cause of cancer death in the worldwide.7,8 Alarmingly, it is increasing sharply in some Asian countries including China.9 MSI has been regarded as one of the three main molecular variations implicated in the development of CRC.10 CRC patients showing dMMR/MSI-H had better stage-adjusted clinical outcome and could benefit differently from a variety of therapies including adjuvant chemotherapy, targeted therapy and immunotherapy.11–13 However, MSI-H stage II CRC patients respond inferiorly to individual 5-fluorouracile (5-FU) based chemotherapy.14 Therefore, it is recommended to detect all CRC patients who undergone surgery for MMR/MSI status testing.
As far as we know, MSI can be determined through PCR-CE method, IHC staining technique can identify presence or absence of MMR proteins,3 or next-generation sequencing (NGS) analysis can detect MMR gene mutation.15 Although determination of MSI by PCR-CE method is recommended by many researchers, IHC analysis of MMR proteins expression is often used instead of clinical practice which chiefly because of its widely used in general laboratories. Besides, there are some problems such as costly, high requirement and time-consuming in NGS analysis. Additionally, markers involved in MSI testing assessed by PCR-CE method also vary in different studies, mainly including Bethesda panel (3 dinucleotide repeats and 2 mononucleotide repeats) recommended by the 1997 National Cancer Institute (NCI)-sponsored MSI workshop and Promega systems (5 mononucleotide repeats) emerged later.16 With conflicting results, some studies have declared that IHC for MMR analysis is an advisable surrogate for PCR in the determination of MSI status, while some reported undesired discordance among these two methods.17,18 Misdiagnosis of dMMR/MSI-H status may be responsible for primary resistance to immunotherapy for CRC patients displaying dMMR or MSI. In addition, the association of dMMR/MSI-H and clinicopathological parameters had been evaluated in previous studies, and with evidence that dMMR/MSI-H is associated with proximal tumor location and poor differentiation.19,20 However, most of these studies had a relatively small number of CRC cases and there were relatively few reports for Chinese patients.
Based on the studies above, we further evaluated the concordance between deficient MMR and MSI testing and their association with clinicopathological features in a large cohort of 738 Chinese patients pathologically diagnosed with CRC. Our study provided reliable evidence for which CRC patients must undergo deficient MMR/MSI testing on the basis of clinicopathological data and offered some valuable suggestions on the selection of detection methods, thus contributing to personalized management of CRC patients.
Materials and Methods
Samples
738 patients who received surgical CRC resections during April 2016 and October 2018 at The Department of Gastrointestinal Surgery, The First Affiliated Hospital of Chongqing Medical University (Chongqing, China) and were then pathologically diagnosed with CRC were enrolled in our study. Patients with insufficient clinicopathological data or specimens were excluded from our study. According to the 8th edition of the American Joint Committee on Cancer (AJCC), the TNM staging was defined.21 Clinicopathological data including age, sex, tumor location, differentiation, histologic type, distant metastasis, proliferation maker Ki-67 protein expression and TNM stage were retrospectively collected from medical records. Formalin-fixed paraffin-embedded (FFPE) tumor tissues and paired peripheral blood specimens of 738 CRC cases were applied to subsequent IHC and MSI detections. Our study was approved by the ethics review board of The First Affiliated Hospital of Chongqing Medical University and informed consent was obtained from all patients. This clinical study was conducted in accordance with the Declaration of Helsinki. The written informed consent, including molecular genetic test and medical record reviews, was obtained from each patient or their next of kin. The data were anonymized for analysis to protect patients’ confidentiality.
Assessment of Deficient MMR by IHC Analysis
FFPE tumor tissue sections of each CRC patients were divided into two parts, one for IHC analysis and the other for MSI testing by PCR-CE method. Briefly, FFPE tumor tissue sections were dewaxed, hydrated and rinsed with phosphate buffer saline (PBS) successively. Endogenous peroxidase activity was inhibited by peroxidase blocker. Primary antibodies which were specific to MSH1, MSH2, MSH6 and PMS2 proteins were added at 4°C overnight, respectively. Following this, the sections were reacted with biotinylated goat anti-mouse IgG antibody and peroxidase-labeled streptavidin successively, and then were detected by streptomycin-avidin-biotin-peroxidase complex (SABC) method, following by color rendering with diaminobenzidine (DAB) and counterstaining with hematoxylin-eosin. Slides dehydration and sealing were finally performed routinely. The aforementioned antibodies were provided by Maixin Biotechnology Development Co. Ltd. (Fuzhou, China). Furthermore, under the condition of positive expression, para-carcinoma tissues including normal epithelial cells, lymphocytes, and mesenchymal cells were used as the internal control. MLH1, MSH2, MSH6 and PMS2 proteins were all located in the nucleus. Deficiency of any products of these four MMR proteins was stated as dMMR, while proficient MMR (pMMR) was determined if all MMR proteins positively expressed.
DNA Extraction and MSI Analysis
In order to obtain higher-purity tumor areas, tumor tissue sections were evaluated by professional pathologist again. DNA was extracted from the tumor-rich areas tissues of each patient’s colorectal tumor and paired peripheral blood specimens as the normal control using a universal FFPE DNA extraction kit according to the manufacturer’s protocol (Amoy Diagnostics Co. Ltd, Xiamen, China). DNA concentration was measured using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific Co. Ltd, Rockford, USA). MSI analysis in CRC cases was performed by PCR-CE method and comparing amplified microsatellite repeats size along with DNA in tumor cells versus paired white blood cells of peripheral blood specimens using microsatellite instability analysis kit (Beijing Microread Gene Technology, Co. Ltd, Beijing, China). DNA from tumor tissues and peripheral blood specimens were detected using a panel of six mononucleotide repeats (BAT25, BAT26, NR21, NR24, NR27 and MONO27) for MSI analysis. Meanwhile, another two pentanucleotide repeats (Penta C, Penta D) and one gender loci (Amel) were also applied to identify tissue mix-up. Each antisense primer of above markers was labeled with a fluorescent dye. The test was carried out according to the manufacturer’s instructions. MSI detection data were analyzed by GeneMapper software (Thermo Fisher Scientific). In comparison with DNA in paired peripheral blood specimens, MSS was defined if none of six markers showed instability in DNA of tumor tissues, MSI-low (MSI-L) if only one marker showed instability and MSI-H if two or more markers demonstrated instability by referring to the NCI standard.
Statistical Analysis
Categorical data were summarized with frequencies. The Pearson chi-square test or Fisher’s exact test was used as appropriate to compare categorical variables. All statistical analyses were evaluated using GraphPad Prism 5 and statistical significance is indicated as p<0.05.
Results
CRC Patient Characteristics
A total of 738 patients pathologically diagnosed with CRC were enrolled in our study. According to clinicopathological features, we found that most of the patients under surgery were older than 50 years old. A total of 174, 183, and 381 patients were diagnosed with left-sided colon cancer, right-sided colon cancer, and rectal cancer, respectively. Moreover, the vast majority of them were well or moderate differentiated, non-mucinous and non-distant metastasis tumors. The data showed that 724 (98.10%) patients had a high expression of Ki-67 (≥20%). CRC was more inclined to occur in TNM stage II (44.44%) and III (35.50%) than other stages (Table 1).
 |
Table 1 Clinicopathological Characteristics of CRC Patients |
Assessment of MMR Proteins Expression Using IHC Method
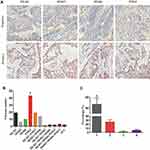
131 of 738 (17.8%) CRC cases with dMMR were observed in our study, representative negative and positive samples of MLH1, MSH2, MSH6 and PMS2 proteins expression assessed by IHC method are shown in Figure 1A. In detail, we found that PMS2 protein loss (43, 32.82%) was the most frequent deficiency among these four proteins, following by the separate deficiency of MLH1 (29, 22.14%), meanwhile, the deficient accident of combined MLH1 with PMS2 (19, 14.50%) or combined MSH2 with MSH6 (14, 10.69%) also played an important role in all types (Figure 1B). In terms of the number of deficiency, only one MMR protein loss (67.10%) was the most part in all CRC patients defined as dMMR (Figure 1C).
Assessment of MSI Status Using PCR-CE Method
Among these tumors, MSI status was established using PCR-CE method. 738 CRC patients were correctly classified in the panel of six mononucleotide repeat markers, and representative examples of MSI phenotypes are presented by Figure 2. 74 of 738 (10.03%) CRC cases with MSI-H were found in our study, of which most (55, 74.32%) showed instability at all six mononucleotide repeat markers. The remainders were composed of only one case with MSI-L and 663 cases with MSS (Figure 3).
Comparison of the Concordance of MMR Proteins Expression and MSI Status
A recent study demonstrated that patients with a dMMR/MSI-H phenotype had a promising response to immunotherapy.22 Hence, it is essential for the reliability in detecting MMR/MSI status. In our study, we found that MSI-H was statistically associated with dMMR (p<0.001; Table 2). Additionally, moderate concordance between IHC method for MMR proteins expression and PCR-CE method for MSI status was observed in our study through Kappa test with a concordance rate of 0.547 (Table 2). Subsequently, we also analyzed the discordant tumors of loss of MMR proteins expression and MSI status as depicted in Figure 4. 69 tumor samples which exhibited a dMMR with IHC but a MSS phenotype with PCR-CE was tested as more commonly MLH1 protein deficiency (59.4%, p=0.029), nevertheless, 62 tumor samples which exhibited a dMMR/MSI-H molecule phenotype were identified as PMS2 protein deficiency mostly (32.2%), although it was not statistically significant (p=0.764). Moreover, it had also to be noted that 12 samples with pMMR were found to display instability at all six mononucleotide repeat markers (data not given).
 |
Table 2 Comparison of MMR Tested by IHC Technique and MSI Status Analyzed by PCR-CE Method in CRC Patients |
Association of MMR Proteins Expression and MSI Status with Clinicopathological Features from CRC Patients
We then associated MMR proteins expression and MSI status with clinicopathological features from CRC patients and performed a pooled analysis including age, sex, tumor location, differentiated degree, distant metastasis, histology type, Ki-67 protein expression and TNM stage. As shown in Table 3, dMMR was more likely to occur in CRC patients under 50 years old (28.24% vs 14.66%, p<0.001). Meanwhile, dMMR was more likely to occur in right-sided colon (p<0.001), poor differentiation tumor tissues (p=0.048), but not associated with distant metastasis, Ki-67 protein expression and TNM stage, consistent with previous literature.22 Similarly, as shown in Table 4, MSI-H occurred in CRC patients under 50 years old (36.49% vs 14.91%, p<0.001). Compared with MSI-L/MSS, MSI-H was associated with right-sided colon (p<0.001), poor differentiation (p<0.001), mucinous type (p=0.037) and TNM stage II (p<0.001), and was independent of distant metastasis and Ki-67 protein expression.
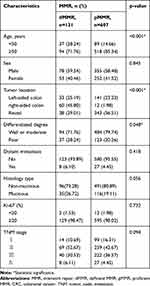 |
Table 3 Association of MMR Proteins Expression with Clinicopathological Characteristics from CRC Patients |
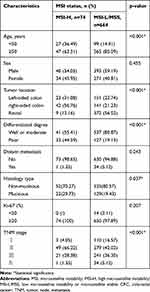 |
Table 4 Association of MSI Status with Clinicopathological Characteristics from CRC Patients |
Discussion
Various nucleotide makers have been proposed to detect MSI status. In 1998, the NCI recommended a panel of five markers including BAT25, BAT26, D2S123, D5S346 and D17S250, and a pattern known as Bethesda panel.23 However, the NCI meeting in 2002 disputed the limitations of three dinucleotide repeats used to assess the microsatellites status including D2S123, D5S346 and D17S250, and also advised another more sensitive and specific system (NR27, NR21, NR24, BAT25 and BAT26).24 According to our results based on the system recommended by NCI meeting in 2002, MSI-H occurred in 74 CRC patients, of which 74.3% showed instability at all six mononucleotide repeats. Youn Jin et al25 presented a very evidential disagreement in the results of the two MSI systems and found higher intratumoral heterogeneity (ITH) originated from dinucleotide repeat markers. Furthermore, mononucleotide repeats were also confirmed to simplify interpretation of the data.26,27 MONO27, a mononucleotide repeat that is out of the system recommended by NCI meeting in 2002, was also applied in our study and demonstrated to be another valuable marker in MSI determination which showed instability in most CRC patients with MSI-H in our study. We further optimized the panel for MSI detection by checking the correspondence between tumor tissues DNA and paired peripheral blood specimens DNA. Microsatellite status was defined as MSI-H, MSI-L and MSS on the basis of above six mononucleotide repeats, MSI-L and MSS were classified as the same subgroup in our study because no obvious differences were observed in clinical manifestations and pathological parameters of these two molecular phenotypes.
dMMR is clinically equivalent to MSI-H, whereas pMMR is the same as MSI-L or MSS.28 According to our results, among the 131 CRC patients with dMMR detected by IHC, PMS2 protein loss was the most commonly observed pattern (60 cases), followed by the MLH1 protein, which is also consistent with previous literature.29 A recent study demonstrated that PMS2 gene could manifest LS phenotype in other ways.30 In addition, our study reported similar numbers of absent MLH1/PMS2 and MSH2/MSH6 proteins, and it may be associated with protein heterodimers, MSH2 dimerizes with MSH6 and MLH1 dimerizes with PMS2 form the functional complex Escherichia coli MutS homologs α (MutSα) and E.coli MutL homologs α (MutLα), respectively.31 We were surprised to find that the combined deficiency of MSH2 and MSH6 proteins was higher than MSH6 protein alone. This is because that MSH2 protein is the prerequisite of their heterodimer, mutation of MSH2 often causes concurrent loss of MSH2 and MSH6 proteins, whereas MSH6 mutation often causes MSH6 protein loss only. Because the function of the secondary protein MSH6 may be compensated by other proteins, such as MSH3.32 Some scholars proposed that proliferating cell nuclear antigen could increase the mismatch-binding specificity of MSH2 and MSH6.33 To our knowledge, IHC technique is economical, of low requirement for experimental instrument, making it a more commonly used method to assess MMR/MSI status in clinic practice. Besides, the variations of MMR genes may be identified indirectly because of loss of MMR proteins staining, providing reference for further determination of targeted DNA sequences. When used in this fashion, however, we must notice that deficiency in specific MMR proteins may result from mutations in a different MMR gene or in other genes associated with CRC.34
Some recent studies declared that IHC technique had virtually equivalent value to PCR method for MSI testing while some questioned.18,32 In our study, both IHC and PCR method were successfully performed and Kappa test showed moderate concordance between these two methods; however, we also observed the bigger discordance, especially in dMMR/MSS groups which were determined by a majority of MLH1 protein deficiency. A study from Yu G et al explained our question35 that germline mutation of MMR was likely to lead to MMR proteins deficiency, but was not demonstrated by PCR method, because these mutations occur in a very early stage of oncogenesis. Furthermore, MLH1 promoter methylation could also generate discordance between MMR proteins deficiency and MSI status.36,37 The discrepancy could be partly explained by variable technical protocols in different laboratories, leading to variations in staining quality and difficulty in interpretation of IHC results. Abdel-Rahman et al also reported that CRC patients with MSH6 mutations did not demonstrate MSI-H by PCR-CE because of a functional redundancy in the MMR system but demonstrated loss of MSH6 staining by IHC technique.38 As we shown, there were 12 samples determined as MSI-H by PCR-CE method but pMMR by IHC method. To our knowledge, over one-third of MLH1 mutations were proved to be missense mutations, which resulted in mutant proteins that were catalytically inactive but antigenically intact, thus making MSI-H by PCR-CE method but pMMR by IHC method.39 Besides, other factors including ITH and even clinic treatment will influence IHC analysis.37,40 Some researchers found that tumor heterogeneity could influence MMR/MSI status.41 Remarkably, a recent study presented by Cohen et al revealed that misdiagnosis of MMR/MSI status was observed if only one method was used and led to primary resistance to immune checkpoint inhibitors in mCRC patients.42 The two assays together are complementary and failure to diagnose would preclude recognition and clinical care. Studies found that assessment of dMMR/MSI-H status had a false positive of 9% in CRC patients included in trials of anti-PD-1.42 Therefore, we actively advocate that both IHC and PCR-CE methods should be routinely tested for MMR/MSI to provide reliable data for clinical treatment decisions, in view of moderate concordance between MMR and MSI testing in our study. Besides, more advanced technology such as NGS technology may have a more thorough assessment.15 Several studies demonstrated that NGS-based method is probably 95.8–100% concordant with PCR-CE testing.43
Previous studies showed that the frequency of MSI-H was ~15%.44 In the present study, of the 738 CRC patients, 10.03% cases were with MSI-H phenotype, a slightly lower than that previously reported, which might be partly ascribed to distinct detection methods and diversity of CRC patients enrolled. Our results showed that dMMR/MSI-H was statistically associated with less than 50 years age, right-sided colon and poor differentiation, which is in keeping with some other publications.19,45,46 Significant difference observed in tumor location suggested that mechanisms of right-sided and left-sided colon cancer might differ at the genetic level. Right-sided cancer occurs mainly due to mutations in tumor-associated genes caused by replication error (RER), while left-sided cancer is mainly related to mutations in oncogenes and tumor suppressor genes through loss of heterozygosity (LOH), indicating that dMMR/MSI-H is mainly involved in the development of right-sided colon cancer.22 Our study showed that dMMR/MSI-H was inclined to poor differentiation, suggesting that CRC with MSI-H is more malignant. However, some studies have reported that CRC patients harboured MSI-H status had a swelling growth pattern and were found to have low growth activity through flow cytometry (FCM), suggesting a benign tendency.47 Hence, the degree of malignancy exhibited by CRC patients with MSI-H in biological behavior need to be further studied. We also found that dMMR/MSI-H was not associated with Ki-67 expression, which is in keeping with the view that the expression of Ki-67 did not correlate with CRC biological behavior and prognosis.48,49 Additionally, our data also showed that MSI-H was significantly associated with TNM stage II. Several studies demonstrated that 5-FU-based adjuvant chemotherapy not only was invalid but it might also have a detrimental impact on survival in stage II MSI-H CRC patients.44 Since MMR/MSI phenotype is related to prognosis and responses to diverse clinic treatments; hence, MMR/MSI testing plays a vital role in personalized management of CRC patients especially in the most prevalent immunotherapy.
Unfortunately, we did not distinguish between sporadic and hereditary CRC patients which were enrolled in our study. More detailed family histories and other genetic analyses including BRAF V600E mutation, hypermethylation of MLH1 promoter and germline mutations of MMR genes should be further considered by other methods. However, we could guide the next plan for hereditary CRC diagnosis and genetic counseling. Hereditary CRC is caused by a germline mutation in one of the MMR genes. When CRC patients with MLH1 loss, we should detect MLH1 hypermethylation and BRAF V600E mutation firstly. These two events are rarely seen in hereditary CRC and therefore may be helpful for determining whether a MSI-H CRC is more likely to be sporadic. The reverse, the other MMR genes mutation should be screened by NGS method.50 In a word, a more sensitive and accurate technique for MMR/MSI assessment will probably be an urgent question and could help more patients to benefit from immunotherapy.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yamamoto H, Imai K. Microsatellite instability: an update. Arch Toxicol. 2015;89(6):899–921. doi:10.1007/s00204-015-1474-0
2. Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi:10.1126/science.aan6733
3. Ryan E, Sheahan K, Creavin B, Mohan HM, Winter DC. The current value of determining the mismatch repair status of colorectal cancer: a rationale for routine testing. Crit Rev Oncol Hematol. 2017;116:38–57. doi:10.1016/j.critrevonc.2017.05.006
4. Zaanan A, Shi Q, Taieb J, et al. Role of deficient DNA mismatch repair status in patients with stage III colon cancer treated with FOLFOX adjuvant chemotherapy: a pooled analysis from 2 randomized clinical trials. JAMA Oncol. 2018;4(3):379–383. doi:10.1001/jamaoncol.2017.2899
5. Kawakami H, Zaanan A, Sinicrope FA. Implications of mismatch repair-deficient status on management of early stage colorectal cancer. J Gastrointest Oncol. 2015;6(6):676–684. doi:10.3978/j.issn.2078-6891.2015.065
6. Bonneville R, Krook MA, Kautto EA, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017;2017.
7. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
8. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
9. Sung JJ, Lau JY, Goh KL, Leung WK. Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol. 2005;6(11):871–876. doi:10.1016/S1470-2045(05)70422-8
10. Devaud N, Gallinger S. Chemotherapy of MMR-deficient colorectal cancer. Fam Cancer. 2013;12(2):301–306. doi:10.1007/s10689-013-9633-z
11. Cortes-Ciriano I, Lee S, Park WY, Kim TM, Park PJ. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun. 2017;8(1):15180. doi:10.1038/ncomms15180
12. Bupathi M, Wu C. Biomarkers for immune therapy in colorectal cancer: mismatch-repair deficiency and others. J Gastrointest Oncol. 2016;7(5):713–720. doi:10.21037/jgo.2016.07.03
13. Sinicrope FA, Okamoto K, Kasi PM, Kawakami H. Molecular Biomarkers in the Personalized Treatment of Colorectal Cancer. Clin Gastroenterol Hepatol. 2016;14(5):651–658. doi:10.1016/j.cgh.2016.02.008
14. Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349(3):247–257. doi:10.1056/NEJMoa022289
15. Middha S, Zhang L, Nafa K, et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol. 2017;2017.
16. Murphy KM, Zhang S, Geiger T, et al. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn. 2006;8(3):305–311. doi:10.2353/jmoldx.2006.050092
17. Do JH, Chang SK, Ahnn JY, et al. Is immunohistochemistry for MLH1 and MSH2 proteins a useful method for detection of microsatellite instability in sporadic colorectal cancer. Korean J Gastroenterol. 2003;42(5):369–376.
18. Qin Y, Liang L, Zheng X, et al. Value of detection of DNA mismatch repair proteins deficiency by immunohistochemistry in predicting tumor microsatellite status. Zhonghua Bing Li Xue Za Zhi. 2015;44(10):704–708.
19. Irabor DO, Oluwasola OA, Ogunbiyi OJ, et al. Microsatellite instability is common in colorectal cancer in native Nigerians. Anticancer Res. 2017;37(5):2649–2654.
20. Kim H, Jen J, Vogelstein B, Hamilton SR. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol. 1994;145(1):148–156.
21. Amin MB, Greene FL, Edge SB, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67(2):93–99. doi:10.3322/caac.21388
22. Domingo E, Ramamoorthy R, Oukrif D, et al. Use of multivariate analysis to suggest a new molecular classification of colorectal cancer. J Pathol. 2013;229(3):441–448. doi:10.1002/path.4139
23. Boland CR, Thibodeau SN, Hamilton SR, et al. A national cancer institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–5257.
24. Campanella NC, Berardinelli GN, Scapulatempo-Neto C, et al. Optimization of a pentaplex panel for MSI analysis without control DNA in a Brazilian population: correlation with ancestry markers. Eur J Hum Genet. 2014;22(7):875–880. doi:10.1038/ejhg.2013.256
25. Choi YJ, Kim MS, An CH, Yoo NJ, Lee SH. Regional bias of intratumoral genetic heterogeneity of nucleotide repeats in colon cancers with microsatellite instability. Pathol Oncol Res. 2014;20(4):965–971. doi:10.1007/s12253-014-9781-y
26. Bacher JW, Flanagan LA, Smalley RL, et al. Development of a fluorescent multiplex assay for detection of MSI-High tumors. Dis Markers. 2004;20(4–5):237–250. doi:10.1155/2004/136734
27. Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123(6):1804–1811. doi:10.1053/gast.2002.37070
28. Hegde M, Ferber M, Mao R, Samowitz W, Ganguly A. ACMG technical standards and guidelines for genetic testing for inherited colorectal cancer (Lynch syndrome, familial adenomatous polyposis, and MYH-associated polyposis). Genet Med. 2014;16(1):101–116. doi:10.1038/gim.2013.166
29. Joehlin-Price AS, Perrino CM, Stephens J, et al. Mismatch repair protein expression in 1049 endometrial carcinomas, associations with body mass index, and other clinicopathologic variables. Gynecol Oncol. 2014;133(1):43–47. doi:10.1016/j.ygyno.2014.01.017
30. Liccardo R, Della Ragione C, Mitilini N, De Rosa M, Izzo P, Duraturo F. Novel variants of unknown significance in the PMS2 gene identified in patients with hereditary colon cancer. Cancer Manag Res. 2019;11:6719–6725. doi:10.2147/CMAR.S167348
31. Dahlin AM, Henriksson ML, Van Guelpen B, et al. Colorectal cancer prognosis depends on T-cell infiltration and molecular characteristics of the tumor. Mod Pathol. 2011;24(5):671–682. doi:10.1038/modpathol.2010.234
32. Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10(4):293–300. doi:10.2353/jmoldx.2008.080031
33. Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet. 2000;26(3):375–378. doi:10.1038/81708
34. Hansen MF, Johansen J, Sylvander AE, et al. Use of multigene-panel identifies pathogenic variants in several CRC-predisposing genes in patients previously tested for Lynch Syndrome. Clin Genet. 2017;92(4):405–414. doi:10.1111/cge.12994
35. Yu G, Wu Y, Wang W, et al. Low-dose decitabine enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by re-modulating the tumor microenvironment. Cell Mol Immunol. 2019;16(4):401–409. doi:10.1038/s41423-018-0026-y
36. Chapusot C, Martin L, Puig PL, et al. What is the best way to assess microsatellite instability status in colorectal cancer? Study on a population base of 462 colorectal cancers. Am J Surg Pathol. 2004;28(12):1553–1559. doi:10.1097/00000478-200412000-00002
37. Lee JH, Cragun D, Thompson Z, et al. Association between IHC and MSI testing to identify mismatch repair-deficient patients with ovarian cancer. Genet Test Mol Biomarkers. 2014;18(4):229–235. doi:10.1089/gtmb.2013.0393
38. Abdel-Rahman WM, Mecklin JP, Peltomaki P. The genetics of HNPCC: application to diagnosis and screening. Crit Rev Oncol Hematol. 2006;58(3):208–220. doi:10.1016/j.critrevonc.2005.11.001
39. Salahshor S, Koelble K, Rubio C, Lindblom A. Microsatellite Instability and hMLH1 and hMSH2 expression analysis in familial and sporadic colorectal cancer. Lab Invest. 2001;81(4):535–541. doi:10.1038/labinvest.3780262
40. Goldstein JB, Wu W, Borras E, et al. Can Microsatellite Status of Colorectal Cancer Be Reliably Assessed after Neoadjuvant Therapy? Clin Cancer Res. 2017;23(17):5246–5254. doi:10.1158/1078-0432.CCR-16-2994
41. Evrard C, Tachon G, Randrian V, Karayan-Tapon L, Tougeron D. Microsatellite instability: diagnosis, heterogeneity, discordance, and clinical impact in colorectal cancer. Cancers. 2019;11(10):1567. doi:10.3390/cancers11101567
42. Cohen R, Hain E, Buhard O, et al. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol. 2019;5(4):551–555. doi:10.1001/jamaoncol.2018.4942
43. Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7(3):746–756. doi:10.1002/cam4.1372
44. Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28(20):3219–3226. doi:10.1200/JCO.2009.27.1825
45. Gong R, He Y, Liu XY, et al. Mutation spectrum of germline cancer susceptibility genes among unselected Chinese colorectal cancer patients. Cancer Manag Res. 2019;11:3721–3739. doi:10.2147/CMAR.S193985
46. Gelsomino F, Barbolini M, Spallanzani A, Pugliese G, Cascinu S. The evolving role of microsatellite instability in colorectal cancer: a review. Cancer Treat Rev. 2016;51:19–26. doi:10.1016/j.ctrv.2016.10.005
47. Gervaz P, Bucher P, Morel P. Two colons-two cancers: paradigm shift and clinical implications. J Surg Oncol. 2004;88(4):261–266. doi:10.1002/jso.20156
48. Allegra CJ, Parr AL, Wold LE, et al. Investigation of the prognostic and predictive value of thymidylate synthase, p53, and Ki-67 in patients with locally advanced colon cancer. J Clin Oncol. 2002;20(7):1735–1743. doi:10.1200/JCO.2002.07.080
49. Cohen T, Prus D, Shia J, et al. Expression of P53, P27 and KI-67 in colorectal cancer patients of various ethnic origins: clinical and tissue microarray based analysis. J Surg Oncol. 2008;97(5):416–422. doi:10.1002/jso.20989
50. Xicola RM, Llor X, Pons E, et al. Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J Natl Cancer Inst. 2007;99(3):244–252. doi:10.1093/jnci/djk033
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.