Back to Journals » OncoTargets and Therapy » Volume 13
Evaluation of C5orf66-AS1 as a Potential Biomarker for Predicting Early Gastric Cancer and Its Role in Gastric Carcinogenesis
Authors Zhou Q, Li H, Jing J , Yuan Y, Sun L
Received 25 November 2019
Accepted for publication 8 March 2020
Published 2 April 2020 Volume 2020:13 Pages 2795—2805
DOI https://doi.org/10.2147/OTT.S239965
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Quan Zhou,1 Hao Li,1 Jingjing Jing,1– 3 Yuan Yuan,1– 3 Liping Sun1– 3
1Tumor Etiology and Screening Department of Cancer Institute, The First Hospital of China Medical University, Shenyang 110001, People’s Republic of China; 2Key Laboratory of Cancer Etiology and Prevention, The First Hospital of China Medical University, Liaoning Provincial Education Department, Shenyang 110001, People’s Republic of China; 3Key Laboratory of Gastrointestinal Cancer Etiology and Prevention, The First Hospital of China Medical University, Liaoning Provincial Department, Shenyang 110001, People’s Republic of China
Correspondence: Liping Sun; Yuan Yuan
Tumor Etiology and Screening Department of Cancer Institute, The First Hospital of China Medical University, 155 Nanjing Street, Shenyang, Liaoning 110001, People’s Republic of China
Tel +86-24-83282153
Email [email protected]; [email protected]
Background: Long non-coding RNAs (lncRNAs) participate in a series of pathological processes in tumorigenesis. Reports show that C5orf66-AS1, an antisense lncRNA, is expressed in various tumors. However, the role of C5orf66-AS1 in gastric cancer (GC) has not been fully clarified. The study focused on the expression patterns and serum level of C5orf66-AS1 in GC to explore its potential application in GC screening and diagnosis. The effects of C5orf66-AS1 on GC cells were also analyzed.
Methods: Tissue and serum samples were used for RNA isolation. Expression levels of C5orf66-AS1 in GC tissues, serum, and cell lines were detected using quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR). CCK-8, transwell, and wound healing assays were performed to determine the effects of C5orf66-AS1 on GC cell behavior.
Results: C5orf66-AS1 expression was downregulated in GC cells compared to that in adjacent normal tissues. Serum C5orf66-AS1 levels were significantly lower in GC patients than in superficial gastritis (GS) and atrophic gastritis (GA) patients. Low serum expression of C5orf66-AS1 was associated with an increased risk of gastric dysplasia (GD) and GC. Receiver operating characteristic curve results showed that the area under curve (AUC) for GC was 0.688, with a sensitivity and specificity of 77.5% and 53.6%, respectively. For the GD + early gastric cancer (ECG) group, the AUC was 0.789, with a sensitivity and specificity of 85.15% and 62.86%, respectively. Correlation analyses of clinicopathological parameters showed that serum C5orf66-AS1 was predominantly associated with Lauren type, TNM stages, pTNM stages, and vessel tumor emboli. Additionally, in vitro overexpression of C5orf66-AS1 in AGS cells inhibited cell proliferation, migration, and invasion.
Conclusion: Decreased expression levels of serum C5orf66-AS1 can be utilized for diagnosis of GC, especially for early diagnosis. The low level of serum C5orf66-AS1 indicated poor biological behavior of tumors in GC patients. In addition, C5orf66-AS1 can inhibit GC cell proliferation, migration, and invasion.
Keywords: gastric cancer, long non-coding RNA, C5orf66-AS1, early diagnosis, cell proliferation, migration and invasion
Introduction
Gastric cancer (GC) is an aggressive malignant disease of gastrointestinal tumor with the second leading cause of cancer-related deaths.1 Early detection and diagnosis are essential for reducing the mortality rate of GC, prompting the requirement of novel efficient biomarkers for diagnosing EGC. It has been widely accepted that besides gastroscopy and mucosal biopsy,2 non-invasive methods, such as Helicobacter pylori (HP) test,3 detection of tumor-associated autoantibody signature,4 serum pepsinogens,5 Gastrin-17 (G-17), nanosensor technology for detection of volatile components in the exhaled breath,6 etc., also play an important role in screening and early diagnosis of gastric cancer.
Currently, the application of non-coding RNAs (circRNAs,7 miRNAs,8 and lncRNAs9) detection in circulating blood for gastric cancer screening has also attracted much attention. Studies have suggested that abnormal expression of lncRNAs may precede that of protein alteration in tumors. lncRNAs are RNA molecules similar to mRNA in structure with transcripts longer than 200 nucleotides (nts) lacking protein-coding ability.9 lncRNAs play a critical role in the genetic,10 epigenetic,11 and post-transcriptional regulation of tumorigenesis.12,13 Comparing with proteins, lncRNAs are generally tissue-specific,14 stable in serum and not easily degraded by RNase.15,16 These characteristics provide a theoretical basis for considering lncRNAs as biomarkers for cancer. It has been reported that lncRNA can participate in series of pathological processes in tumorigenesis, for instance, inflammatory response,17 autophagy,18 pyroptosis,19 metabolic reprogramming,18 as well as crosstalk between immune cells and tumor cells20 and immunotherapy resistance.21 The extensive functions of lncRNAs suggest extremely valuable for research.
C5orf66-AS1 (also named CTC-276P9.1 or Epist) is an antisense lncRNA present at the first intron region of C5ORF66, which is located on chromosome 5 (chr5: 135, 038, 831-135, 040, 047; size: 1, 217), containing two exons. A few studies reported that downregulation of C5orf66-AS1 was associated with pituitary null cell adenomas,17 oral squamous cell carcinoma,18,19 head and neck squamous cell carcinomas,20 esophageal squamous cell carcinoma,21 and bladder cancer;22 however, it was upregulated in cervical cancer.23 Aberrant hypermethylation-mediated downregulation of C5orf66-AS1 might serve as a potential prognostic biomarker for cardia adenocarcinoma.24 In our former study, C5orf66-AS1 was identified by mRNA-lncRNA co-expression profiling microarray screening and was subsequently verified by The Cancer Genome Atlas (TCGA) databases (data not provided). However, the expression patterns, relationships with clinicopathological parameters of GC patients, and biological functions of C5orf66-AS1 are still unclear.
In the current study, we determined C5orf66-AS1 expression levels in both tissues and serum to illustrate expression characteristics and explored the potential of C5orf66-AS1 for GC diagnosis. The relationship between C5orf66-AS1 expression levels in serum and clinicopathological parameters was also analyzed. Moreover, we investigated the effects of C5orf66-AS1 on the proliferation, migration, and invasion of GC cells in vitro, in an attempt to explain the role of C5orf66-AS1 in gastric carcinogenesis.
Materials and Methods
Patients and Specimens
Seventy-six matched samples of GC and tumor-adjacent non-cancerous tissues were collected from patients with GC who underwent surgical resection without preoperative physical or chemical therapies or blood transfusion before surgery in the First Hospital of China Medical University, Shenyang, China from 2013 to 2017. Patients included 50 males and 26 females with average ages of 63 and 58 years, respectively. Tissue samples were placed in RNAlater solution (RNAlater™ Stabilization Solution, Thermo Fisher Scientific, Waltham, MA, USA) immediately after surgery and were subsequently frozen at –80°C until RNA extraction. Detailed clinical data were collected from the medical records of the hospital, including gender, age, HP infection, smoking, alcohol consumption, Lauren type, TNM classification, pTNM classification, lymph node invasion, tumor-infiltrating lymphocytes, vessel tumor emboli, perineural invasion, and extranodal tumor deposits.
A total of 478 serum samples were collected from The First Hospital of China Medical University from 2013 to 2017, and the Zhuanghe Gastric Diseases Screening Program between 2008 and 2017. The enrolled subjects comprised 134 GS, 102 GA, 42 GD, 59 EGC, and 141 advanced gastric cancer (AGC) patients. Three milliliters of peripheral blood were extracted, centrifuged at 3500 × g for 10 min (within 20 min of collection), and all serum samples were frozen at –80°C until RNA extraction.
The current study was approved by the Human Ethics Review Committee of the First Hospital of China Medical University (Shenyang, China). Each participant in the study signed an informed consent.
Total RNA Extraction and Reverse Transcription
Total RNA was extracted with TRIzol Reagent (TaKaRa, Dalian, China) from collected tissues, and serum was processed using a TIANamp Virus RNA Kit (Tiangen Biotech Co. Ltd., Beijing, China). All procedures were performed according to the manufacturers' instructions. The quality of the total RNA from the tissues was detected at an A260/A280 ratio by NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, MA, USA) and only samples with a ratio between 1.8 and 2.0 were used. Total RNA was converted into complementary DNA using a PrimeScript RT Master Mix (TaKaRa, Dalian, China) according to standard protocols.
Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
The expression of C5orf66-AS1 and an internal-control gene (beta-actin) in tissue and serum samples were detected by qRT-PCR, using SYBR PREMIX EX Taq II (TaKaRa, Dalian, China). The assay was performed on a Mastercycler ep realplex (Eppendorf, Hamburg, Germany). Primers for C5orf66-AS1 were as follows: forward 5′- GGTCGGGCTTTTTCTTCCCA-3′ and reverse 5′- GCCGCGGGAATGTCTTTATT-3′. Primers for beta-actin were as follows: forward 5′- ATGTGGCCGAGGACTTTGATT-3′ and reverse 5′- AGTGGGGTGGCTTTTAGGATG-3′. The conditions of thermal cycling were 30 s at 95°C, 40 cycles at 95°C for 10 s, 57°C for 20 s, and 72°C for 30 s. After all these steps, a melting curve was produced. Melting curve analysis was used to verify specificity and to exclude non-specific products and primer dimers. No-template controls were included in each experiment and duplicate reactions were performed.
With the 2-ΔCt method, the relative quantification of C5orf66-AS1 expression was calculated. In each sample, we normalized the expression levels of C5orf66-AS1 to those of beta-actin in each sample using the formula: ΔCt (delta Ct) = Ct target – Ct beta-actin. Relative expression levels were expressed as 2-ΔCt based on ΔCt values.
Cell Culture
Six human gastric adenocarcinoma cell lines, including BGC823, MGC803, SGC7901, AGS, MKN28, and MKN45 and one normal gastric epithelium cell line (GES-1) were obtained from the Department of Cell Genetics at the Beijing Institute for Cancer Research (Beijing, China). BGC823, MGC803, SGC7901, MKN28, MKN45, and GES-1 cells were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) and the AGS cell line was cultured in F12 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Beijing Zhong Shan-Golden Bridge Biological Technology Co., Beijing, China) and 1% penicillin-streptomycin solution (Solarbio, Beijing, China) at 37°C in 5% CO2.
Plasmids and Cell Transfection
The full-length human C5orf66-AS1 transcript 1 was synthesized and cloned into a GV144 vector (Genechem, Shanghai, China), GV144-C5orf66-AS1, while an empty GV144 vector was used as a control. AGS cells (2.5 × 105) were seeded on a 6-well culture plate and cultured until they reached 70–80% confluency. The GV144-C5orf66-AS1 and empty vector were transfected into pre-seeded cells, using jetPRIME transfection reagent (Polyplus Transfection, Illkirch, France) following the instructions of the manufacturer. Cells were cultured for 24 h, and transfection efficiency was identified using a universal microscope (Olympus, Japan), and overexpression efficiency was determined using qRT-PCR.
Cell Proliferation Assay
Briefly, cells were seeded into 96-well plates with a total of 2 × 103 per well containing 100 μL medium. The plates were incubated at 37°C in 5% CO2 for 24, 48, 72, 96 and 120 h. 10 μL of Cell Count Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) solution were added to each well, and the plates were incubated for 2 h. The absorbance was determined at 450 nm using a 96-well Multimode Plate Reader (BioStack Ready, PowerWave XS, BioTek Instruments, Winooski, VT, USA). Each experiment was conducted with three biological replicates.
Transwell Migration and Invasion Assay
In the migration assay, 5 × 104 cells suspended in 1% serum medium, were added into the upper chamber (8 μm pore size; Corning Inc., Corning, NY, USA). The invasion assay was performed using 2 × 105 cells placed on a Matrigel-coated membrane (BD Biosciences, San Jose, CA, USA) to form a matrix barrier. Six hundred milliliters of F12 medium containing 10% fetal bovine serum was used for each lower chamber. After incubation at 37°C for 24 h and 48 h, samples were washed with PBS and fixed in 4% Paraformaldehyde Fix Solution and stained with 0.1% crystal violet. Under a microscope, numbers were counted in four randomly selected regions (200×) (Nikon Eclipse Ti-S, Nikon, Tokyo, Japan). The experiment was conducted with three replicates.
The migration process was evaluated using a wound-healing assay. In brief, 1 × 106 per well AGS cells were seeded into 6-well culture plates, which were incubated overnight. Wounds were made by scratching cell monolayers with a tip. Cells were cultured in FBS-free media and photographed at 0 and 24 hrs, respectively. To determine the migration capacity, the scratch area was measured.
Statistical Analyses
Student’s t-test was used to compare the differences in C5orf66-AS1 expression between GC tissues and the matched non-tumor tissues. ANOVA was used to compare differences in serum C5orf66-AS1 among GS, GA, GD, and GC. Multinomial logistic regression analysis was conducted to calculate the odds ratios (ORs) and their 95% confidence intervals (95% CI) to estimate the associations between serum C5orf66-AS1 and the risk of stomach diseases. The receiver operating characteristic (ROC) curve and the area under curve (AUC) were used for diagnostic effectiveness analysis. All statistical analyses were carried out using SPSS 23.0 software (SPSS, Chicago, IL, USA). P value less than 0.05 is considered as statistical significance.
Results
C5orf66-AS1 Is Lower in GC Tissues
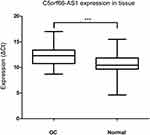
The expression of C5orf66-AS1 was significantly lower in GC tissues (2-ΔCt, 0.0012 ± 0.005 vs 0.005 ± 0.014; P=0.038) than in adjacent normal tissues (Figure 1). In paired tissues, C5orf66-AS1 was downregulated (2-ΔΔCt fold change<1.0) in 61 out of 76 GC tissues (80.3%), and the average expression of C5orf66-AS1 in adjacent normal tissues was 4.17 times higher than that in GC.
Low Serum Expression of C5orf66-AS1 Increased the Risk of GD and GC
We further explored C5orf66-AS1 expression levels in serum in various groups, including GS, GA, GD, total GC, EGC, and AGC. Compared with GS, C5orf66-AS1 expression was significantly decreased in GC serum, especially in GD and EGC (P<0.001) (Figure 2, Table 1).
 |
Table 1 Relationship Between Serum C5orf66-AS1 Level and the Risk of Gastric Diseases |
According to the cutoff value for the GC + GD group, the expression level of C5orf66-AS1 could be divided into two groups: a high expression group (HEG) and a low expression group (LEG). Compared with the HEG group, the risk of GD (OR 0.037, 95% CI = 0.011–0.128, P<0.001) and GC (OR 0.195, 95% CI = 0.121–0.315, P<0.001), including EGC (OR 0.083, 95% CI = 0.036–0.191, P<0.001) and AGC (OR 0.259, 95% CI = 0.156–0.429, P<0.001), significantly increased in the LEG group. However, there was no obvious correlation between C5orf66-AS1 expression and the risk of GA (OR 1.027, 95% CI = 0.589–1.79, P = 0.622) (Table 1).
Serum C5orf66-AS1 Has Good Diagnostic Value for GC, Especially for EGC
We used ROC curve to evaluate the diagnostic efficacy of C5orf66-AS1. For distinguishing GC and non-GC, the AUC reached 0.688 (95% CI = 0.644–0.729; P<0.001; Figure 3A); sensitivity and specificity were 77.5% and 53.6%, respectively, and the cutoff value was 0.134. To determine the diagnostic performance for EGC and AGC, we analyzed them separately. For diagnosing EGC, the AUC reached 0.749 (95% CI = 0.708–0.788; P<0.001) (Figure 3B), with a cutoff value of 0.128, sensitivity of 94.92%, and specificity of 48.21%. Considering that the expression of C5orf66-AS1 began to decline in the GD group, we combined GD with GC or EGC to determine the diagnostic performance. The AUC of the GC + GD group was 0.768 (95% CI = 0.727–0.805, P<0.001) (Figure 3C), with a cutoff value of 0.12. The AUC of the GD + EGC group was 0.789 (95% CI = 0.750–0.825, P<0.001) with a cutoff value of 0.12 (Figure 3D, Table 2).
 |
Table 2 Diagnostic Efficacy of Serum C5orf66-AS1 in Gastric Cancer |
Correlation Between Serum C5orf66-AS1 Levels and Clinical Characteristics of GC Patients
We examined the correlation between serum C5orf66-AS1 levels and clinicopathological parameters of GC patients. Serum C5orf66-AS1 in GC with diffuse type or with vessel tumor emboli was significantly higher than that with intestinal type (P<0.001) or without vessel tumor emboli (P=0.002). In addition, the level of serum C5orf66-AS1 in the advanced stage was significantly higher than that in early stage (P=0.021, P<0.001), whether in TNM stage or pTNM stage (P<0.001). All characteristics are shown in Table 3.
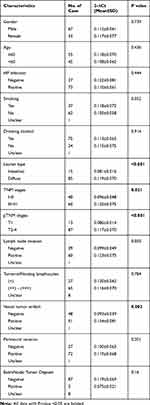 |
Table 3 The Correlation Between Serum C5orf66-AS1 Level and the Clinicopathological Characteristics of GC Patients |
Overexpression of C5orf66-AS1 Suppressed GC Cell Proliferation and Inhibited GC Cell Migration and Invasion in vitro
We detected the basal expression levels of C5orf66-AS1 in the gastric immortalized cell GES-1 and six other GC cell lines, including 7901, MKN45, 823, 803, HGC27, and AGS. AGS, the cell line with the lowest expression of C5orf66-AS1, was selected for the subsequent transfection experiment (Figure 4A). The transfection efficiency was over 80% compared with the original AGS cells, and there was a 50,000–60,000 fold increase in the expression of C5orf66-AS1 in AGS (Figure 4B, C). CCK-8 assay showed that cell growth was significantly suppressed in AGS cells transfected with GV144-C5orf66-AS1, compared with the respective controls (Figure 5).
 |
Figure 5 CCK-8 assay. Cell proliferation was assessed daily for 5 days using a CCK-8 assay. C5orf66-AS1 transfection reduces cell viability in AGS cells, *P<0.05. |
To further explore whether C5orf66-AS1 played a direct functional role in promoting cell invasion in GC, cancer cell invasion was evaluated through transwell matrigel assay. As shown in Figure 6 overexpression of C5orf66-AS1 suppressed the migration of AGS cells compared with the control and original AGS cells; invasion of AGS cells was reduced following overexpression of C5orf66-AS1. Similarly, the wound-healing assay also indicated that C5orf66-AS1 inhibited cell migration and invasion ability (Figure 7). These results indicated that C5orf66-AS1 could suppress the migratory and invasive GC cell phenotypes.
Discussion
In our previous study, C5orf66-AS1, a novel lncRNA that was abnormally expressed in GC, was found by mRNA-lncRNA co-expression microarray screening and TCGA data verification. Based on this result, the present study further illustrated the expression characteristics of C5orf66-AS1 in stomach tissues and serum. The expression levels of C5orf66-AS1 were significantly downregulated in GC tissues, suggesting that it might be involved in GC as a dysregulated tumor suppressor gene. These results provided important support for further study of its application value for the diagnosis of gastric cancer. Moreover, C5orf66-AS1 can inhibit GC cell proliferation, migration, and invasion, implying that C5orf66-AS1 is a crucial factor in gastric carcinogenesis.
lncRNAs are stable in peripheral blood, while abnormal changes in expression of their transcripts may occur earlier than changes in protein, indicating that lncRNAs could be potential diagnostic markers of tumors. The Correa model shows that GC is a multi-stage process, with multiple precancerous stages, including atrophic gastritis, intestinal metaplasia, and dysplasia, which eventually develop into GC.25 It is important to identify precancerous lesions because achieving an early diagnosis can improve the survival rate of GC patients. In this study, we examined the expression levels of C5orf66-AS1 in the serum of GS, GA, GD, and GC patients (Figure 2). Serum C5orf66-AS1 levels were lower in the GD and GC groups (especially in the GD and EGC groups) than in the GS and GA groups (P<0.001). The expression levels of C5orf66-AS1 were significantly lower in these groups, suggesting that C5orf66-AS1 could serve as a potential biomarker in GC early diagnosis.
In further diagnostic efficacy analysis, C5orf66-AS1 showed excellent diagnostic performance for EGC and its AUC for diagnosis of GD + EGC was 0.789, and the sensitivity and specificity reached 85.15% and 62.86%, respectively (Table 2). The AUC for diagnosing EGC was 0.749, and the sensitivity and specificity were 94.92% and 48.21%, respectively (Figure 3 and Table 2). As far as we know, only a few studies have screened EGC by detecting non-invasive molecular expression levels in circulating blood, such as non-coding RNAs (miRNAs, lncRNAs) and serum autoantibodies. miRNA-199a-3p, with high diagnostic efficiency, produced an AUC of 0.818 for diagnosing EGC, and the sensitivity and specificity were 0.76 and 0.74, respectively.26 The AUC for EGC of XIST and RRP1B were reported to be 0.733 and 0.753, and the sensitivity and specificity were 0.846 and 0.590, and 0.859 and 0.564, respectively.27,28 The specificity of serum p53 antibody for EGC diagnosis was as high as 99.15%, but the sensitivity was as low as 12.35%.29 The sensitivity and specificity of serum pepsinogen and gastrin 17 in the diagnosis of EGC were 83% and 68%, respectively.30 Compared with other indicators, therefore, C5orf66-AS1 showed similar or better diagnostic ability, with higher sensitivity for EGC than most other lncRNAs. Indeed, our results provide evidence that serum C5orf66-AS1 levels can be used to distinguish EGC patients from controls. However, expanding the samples of different phases for further verification or prospective observational studies is necessary. In addition, combining other emerging indicators or classic indicators to improve the specificity of diagnosis is also necessary.
The relationship between C5orf66-AS1 expression and the progression of gastric disease was a U-shaped curve rather than a linear correlation. EGC was at the lowest point of the curve. The results of Lauren type, TNM stages, and pTNM stages suggested that C5orf66-AS1 expression levels were lower in the early stages than in the advanced stages (Table 3). This result is in line with the lowest C5orf66-AS1 expression occurring in EGC. Currently, due to a lack of knowledge of the mechanism of C5orf66-AS1 in tumors, there is no reasonable evidence to explain the U-shaped changes in lncRNA with disease progression. As regulatory factors, lncRNAs rely on their combined target gene performing their function. C5orf66-AS1 expression is decreased in various tumors, and low expression in oral squamous cell carcinoma leads to proliferation and other biological behaviors caused by negative regulation of CYC1. In contrast, C5orf66-AS1 is overexpressed in cervical cancer, and acts through ceRNA to increase RING1 expression, thereby promoting malignant behavior. These contrasting results may help to explain the inconsistent expression of C5orf66-AS1 in early and advanced GC.
C5orf66-AS1 inhibited the proliferation, invasion, and metastasis of GC cells, suggesting that C5orf66-AS1 might act as a tumor suppressor gene in GC. At present, the role of C5orf66-AS1 in the development of malignant tumors is not consistent. Guo et al21 reported that C5orf66-AS1 was up-regulated in esophageal cancer cells, the expression of proliferation-related indicators Ki-67 and PCNA was downregulated, the expression of EMT-related proteins N-cadherin and Vimentin was downregulated, and the E-cadherin involved in the invasion and metastasis pathway was upregulated, suggesting that C5orf66-AS1 inhibited the proliferation and metastasis of esophageal cancer cells.21 Lu et al19 reported that CYC1 was negatively regulated by C5orf66-AS1 in oral squamous cell carcinoma, and the biological behavior (invasion, metastasis, proliferation, and apoptosis) could be reversed by knocking down CYC1. Rui et al23 reported that C5orf66-AS1 could promote the proliferation, invasion, and metastasis of cervical cancer cells and inhibit the apoptosis of cervical cancer cells by competing with RING1 in combination with miR-637. Therefore, it acted as a proto-oncogene to promote the development of cervical cancer.23 C5orf66-AS1, as a non-coding RNA, cannot directly exert biological functions but affects the occurrence and development of tumors by regulating different target genes, which may be the reason for its varying biological effects in different tumors. Therefore, finding the target genes of C5orf66-AS1 in GC is crucial for analyzing its role in the process of GC.
Our study has two main limitations. First, we included both EGC and AGC patients for comprehensively diagnostic efficacy analysis. However, EGC cases were still limited, and AGC cases were not evaluated as Phase II and III. In the future, we will define and stabilize the diagnostic efficacy of C5orf66-AS1 for gastric cancer by expanding the sample size of different clinical stages for validation studies. Second, because of the low level of C5orf66-AS1 expression in GC tissues and cells, we just considered overexpressed transiently transfected cells for functional research in the present study, while the controlled functional studies in the reverse direction were not performed.
In conclusion, our results indicate that C5orf66-AS1 may primarily be a tumor suppressor gene. C5orf66-AS1 could suppress the proliferation and migration of cancer cells. Low serum expression of C5orf66-AS1 could be considered for early prediction of GC. Besides, the expression level of serum C5orf66-AS1 indicated poor biological behavior of tumors in GC patients. These results provide a better understanding of the role of C5orf66-AS1 in GC and present a new biomarker for the diagnosis of EGC.
Acknowledgments
This study was supported by grants from The National Key R&D Program (no. 2016YFC1303202).
Author Contributions
Liping Sun contributed to study design and revising the manuscript. Yuan Yuan contributed to experimental instruction and revising the manuscript. Quan Zhou contributed to performing the experiment, data interpretation and drafting manuscript. Hao Li contributed to data interpretation and statistical analysis. Jingjing Jing contributed to sample collection and sample preparation. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts interest in this study.
References
1. Sitarz R, Skierucha M, Mielko J, Offerhaus GJA, Maciejewski R, Polkowski WP. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248. doi:10.2147/CMAR.S149619
2. Pasechnikov V, Chukov S, Fedorov E, Kikuste I, Leja M. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol. 2014;20(38):13842–13862. doi:10.3748/wjg.v20.i38.13842
3. Correa P, Fox J, Fontham E, et al. Helicobacter pylori and gastric carcinoma. Serum antibody prevalence in populations with contrasting cancer risks. Cancer. 1990;66(12):2569–2574. doi:10.1002/1097-0142(19901215)66:12<2569::AID-CNCR2820661220>3.0.CO;2-I
4. Zayakin P, Ancans G, Silina K, et al. Tumor-associated autoantibody signature for the early detection of gastric cancer. Int J Cancer. 2013;132(1):137–147. doi:10.1002/ijc.v132.1
5. Hattori Y, Tashiro H, Kawamoto T, Kodama Y. Sensitivity and specificity of mass screening for gastric cancer using the measurement of serum pepsinogens. Jpn J Cancer Res. 1995;86(12):1210–1215. doi:10.1111/j.1349-7006.1995.tb03317.x
6. Broza YY, Khatib S, Gharra A, et al. Screening for gastric cancer using exhaled breath samples. Br J Surg. 2019;106(9):1122–1125. doi:10.1002/bjs.11294
7. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi:10.1016/j.cca.2015.02.018
8. Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: current insights and future perspectives. World J Gastroenterol. 2018;24(30):3313–3329. doi:10.3748/wjg.v24.i30.3313
9. Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25–64.
10. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi:10.1146/annurev-biochem-051410-092902
11. Amirkhah R, Naderi-meshkin H, Schmitz U, Shah JS, Dunne PD. The intricate interplay between epigenetic events, alternative splicing and noncoding RNA deregulation in colorectal cancer. Cells. 2019;8(8):929. doi:10.3390/cells8080929
12. Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15(3):177–186. doi:10.1016/j.gpb.2016.12.005
13. Wang W, Xie Y, Chen F, et al. LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J Gastroenterol. 2019;25(29):3972–3984. doi:10.3748/wjg.v25.i29.3972
14. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi:10.1101/gr.132159.111
15. Yang Z, Guo X, Li G, Shi Y, Li L. Long noncoding RNAs as potential biomarkers in gastric cancer: opportunities and challenges. Cancer Lett. 2016;371(1):62–70. doi:10.1016/j.canlet.2015.11.011
16. Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom Clin appl. 2015;9(3–4):358–367. doi:10.1002/prca.201400114
17. Yu G, Li C, Xie W, et al. Long non-coding RNA C5orf66-AS1 is downregulated in pituitary null cell adenomas and is associated with their invasiveness. Oncol Rep. 2017;38(2):1140–1148. doi:10.3892/or.2017.5739
18. Feng L, Houck JR, Lohavanichbutr P, Chen C. Transcriptome analysis reveals differentially expressed lncRNAs between oral squamous cell carcinoma and healthy oral mucosa. Oncotarget. 2017;8(19):31521–31531. doi:10.18632/oncotarget.16358
19. Lu T, Liu H, You G. Long non-coding RNA C5orf66-AS1 prevents oral squamous cell carcinoma through inhibiting cell growth and metastasis. Int J Mol Med. 2018;42(6):3291–3299. doi:10.3892/ijmm.2018.3913
20. Sailer V, Charpentier A, Dietrich J, et al. Intragenic DNA methylation of PITX1 and the adjacent long non-coding RNA C5orf66-AS1 are prognostic biomarkers in patients with head and neck squamous cell carcinomas. PLoS One. 2018;13(2):e0192742.
21. Guo W, Liu S, Dong Z, et al. Aberrant methylation-mediated silencing of lncRNA CTC-276P9.1 is associated with malignant progression of esophageal squamous cell carcinoma. Clin Exp Metastasis. 2018;35(1–2):53–68. doi:10.1007/s10585-018-9881-2
22. Zhu YP, Bian XJ, Ye DW, et al. Long noncoding RNA expression signatures of bladder cancer revealed by microarray. Oncol Lett. 2014;7(4):1197–1202. doi:10.3892/ol.2014.1843
23. Rui X, Xu Y, Jiang X, Ye W, Huang Y, Jiang J. Long non-coding RNA C5orf66-AS1 promotes cell proliferation in cervical cancer by targeting miR-637/RING1 axis. Cell Death Dis. 2018;9(12):1175. doi:10.1038/s41419-018-1228-z
24. Guo W, Lv P, Liu S, et al. Aberrant methylation-mediated downregulation of long noncoding RNA C5orf66-AS1 promotes the development of gastric cardia adenocarcinoma. Mol Carcinog. 2018;57(7):854–865. doi:10.1002/mc.v57.7
25. de Vries AC, van Grieken NC, Looman CW, et al. Gastric cancer risk in patients with premalignant gastric lesions: a nationwide cohort study in the Netherlands. Gastroenterology. 2008;134(4):945–952. doi:10.1053/j.gastro.2008.01.071
26. Li C, Li JF, Cai Q, et al. MiRNA-199a-3p: a potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013;108(2):89–92. doi:10.1002/jso.23358
27. Lu Q, Yu T, Ou X, Cao D, Xie T, Chen X. Potential lncRNA diagnostic biomarkers for early gastric cancer. Mol Med Rep. 2017;16(6):9545–9552. doi:10.3892/mmr.2017.7770
28. Cui Z, Chen Y, Xiao Z, et al. Long noncoding RNAs as auxiliary biomarkers for gastric cancer screening: a pooled analysis of individual studies. Oncotarget. 2016;7(18):25791–25800. doi:10.18632/oncotarget.8268
29. Werner S, Chen H, Tao S, Brenner H. Systematic review: serum autoantibodies in the early detection of gastric cancer. Int J Cancer. 2015;136(10):2243–2252. doi:10.1002/ijc.v136.10
30. Shiotani A, Iishi H, Uedo N, et al. Histologic and serum risk markers for noncardia early gastric cancer. Int J Cancer. 2005;115(3):463–469. doi:10.1002/(ISSN)1097-0215
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.