Back to Journals » Infection and Drug Resistance » Volume 11
Evaluation of blood culture broths with lysis buffer to directly identify specific pathogens by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry methods
Authors Hariu M, Watanabe Y , Oikawa N, Manaka T, Seki M
Received 24 March 2018
Accepted for publication 12 June 2018
Published 24 September 2018 Volume 2018:11 Pages 1573—1579
DOI https://doi.org/10.2147/IDR.S169197
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Joachim Wink
Maya Hariu,1,2 Yuji Watanabe,1,2 Nozomi Oikawa,1,2 Takafumi Manaka,1,2 Masafumi Seki1
1Division of Infectious Diseases and Infection Control, Tohoku Medical and Pharmaceutical University Hospital, Sendai City, Miyagi, Japan; 2Department of Laboratory Medicine, Laboratory for Clinical Microbiology, Tohoku Medical and Pharmaceutical University Hospital, Sendai City, Miyagi, Japan
Background: Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is now widely used to detect pathogens in clinical settings in Japan.
Method: Here, we report the effects of adding lysis buffer in the MALDI-TOF MS method to directly detect bacteria from 3 blood culture systems and compare their detection efficiencies for each pathogen.
Results: Blood culture broths from BD, bioMérieux, and Oxoid showed similar detection efficiencies without lysis buffer use and Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa were efficiently detected in all broths when lysis buffer was used. However, Streptococcus pneumoniae was not detected in BD broth when lysis buffer was added. Furthermore, Haemophilus influenzae and Bacteroides fragilis were not detected in all 3 systems when lysis buffer was used.
Conclusion: Optimization of blood culture system and lysis buffer is necessary according to each pathogen for direct identification by MALDI-TOF MS methods.
Keywords: blood cultures broth, lysis buffer, Escherichia coli, Haemophilus influenzae, Streptococcus pneumoniae, time-of-flight mass spectrometry
Introduction
Sepsis often presents as multiple-organ dysfunction, and bacteremia is typically diagnosed by microbiological tests, including blood cultures (BCs).1 However, the pathogens in the blood are detected in only 4%–12% of all BCs, and identification of the pathogens by BC usually take 2–3 days.1,2 Therefore, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been recently applied because it allows the identification of most pathogenic bacteria and fungus grown in BC bottles directly within a few minutes and has been proven as being efficient and reproducible.3–5
A red blood cell (RBC) lysis step is usually needed to detect bacteria clearly because RBCs avoid efficient identification of the pathogenic bacteria, and some lysis buffers were used to identify the pathogens efficiently from BC bottles before MALDI-TOF MS processes. However, among these lysis buffers, some of them were found to lyse not only RBCs but also the targeted pathogenic bacteria.
Therefore, in this study, we investigated the effects of adding lysis buffer in the process of direct pathogen detection and found some bacteria could not be detected in certain BC systems.
Methods
Samples
Blood specimens from adult patients were systematically collected and cultured using the BD BACTEC system (BD, Franklin Lakes, NJ, USA), bioMérieux BacT/ALERT system (bioMérieux, Marcy l’Etoile, France), and Oxoid Signal Blood Culture system (Oxoid, Newport, England), including both anaerobic and aerobic resins. Bottles containing 8–10 mL of whole blood were incubated in MIR-553 instruments (Sanyo, Tokyo, Japan) for 5–7 days for the standard incubation protocol or for fewer days if an earlier positive signal was detected.
Direct identification of bacteria by MALDI-TOF MS from both positive aerobic and anaerobic bottles (Bruker Daltonics, Bremen, Germany) was performed serially in parallel with the routine protocol at the clinical laboratory of Tohoku Medical and Pharmaceutical University Hospital; these tests were performed from July 2014 to December 2017.3,6 After subculturing on agar plates, any positive BC was simultaneously assessed for bacterial identification using conventional procedures, including MicroScan Walkaway panels (Beckman Coulter, Jersey City, NJ, USA), and, if necessary, complementary individual biochemical or enzymatic tests. We performed each experiment 5–10 times. The spectral scores >1.700–1.999 and >2.000 indicated identification to genus and species level, respectively.
This study was approved by the Committee for Clinical Scientific Research of Tohoku Medical and Pharmaceutical University Hospital on July 08 and December 10, 2015 (Nos. ID2015-2-008 and ID2015-2-037, respectively), and all patients provided written informed consent for use of their blood specimens although BC samples were specifically isolated for part of the routine hospital laboratory procedure.
Separation of blood cells and bacteria, protein extraction protocol, and bacterial identification
We used the MALDI Sepsityper kit (Bruker Daltonics). In brief, approximately 1 mL of cultured sample was recovered from a given positive BC bottle, transferred to a microfuge tube, and combined with or without 200 µL lysis buffer (Sepsityper kit). After mixing by vortexing for 10 seconds, tubes were centrifuged at 10,000 g for 2 minutes at room temperature, and supernatants were removed. We then resuspended each pellet by pipetting with 1 mL washing buffer before recentrifuging the samples for 1 minute at 10,000 g. After removing the supernatant, the bacterial pellet was treated according to the standard ethanol/formic acid protein extraction protocol for MALDI-TOF MS identification according to the MBT Sepsityper kit instruction manual.
Preparation of bacteria for MALDI-TOF MS analysis
To examine the ability of MALDI-TOF MS to distinguish samples with or without lysis buffer, we prepared several types of bacterial BC samples and then performed BC broth analysis as described above. The selected bacteria were Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Streptococcus pneumoniae, Haemophilus influenza, and Bacteroides fragilis.
Results
Detection of E. coli, K. pneumoniae, and P. aeruginosa from BC by MALDI-TOF MS with or without lysis buffer
For this comparison of MALDI-TOF MS results with those obtained by routine techniques from clinical samples, E. coli was the one detected most frequently.6
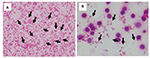
Furthermore, E. coli was detected more clearly when lysis buffer was used (Figure 1). The mean score values of MALDI-TOF MS with or without lysis buffer were 2.315 and 1.779, respectively (Figure 2). This tendency that bacilli were more clearly detected with the addition of lysis buffer was similar among all tested BC systems (Table 1).
Furthermore, K. pneumoniae detection rates were similar to E. coli rates; bacilli were detected from all 3 BC systems with or without lysis buffer (Table 1). In addition, if we used the pellet for MALDI-TOF MS analysis, bacilli were detected more clearly because the concentration and number of bacilli were increased compared with routine samples from suspended pellets.
For P. aeruginosa, we could detect bacilli in all 3 BC systems with lysis buffer but not from bioMérieux BC bottle without lysis buffer because RBCs were mixed if we used suspended pellets (Table 1).
Detection of S. pneumoniae from BC by MALDI-TOF MS with or without lysis buffer
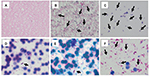
Detection of S. pneumoniae differed from results for Gram-negative pathogens, E. coli, K. pneumoniae, and P. aeruginosa. S. pneumoniae was more clearly detected from bioMérieux and Oxoid BCs when lysis buffer was used. In contrast, in BD BC, bacilli disappeared when lysis buffer was added (Figure 3). S. pneumoniae could not be detected when lysis buffer was not used because RBCs were mixed in BD and bioMérieux BC bottles (Table 1).
Detection of H. influenzae and B. fragilis from BC by MALDI-TOF MS with or without lysis buffer
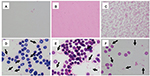
In H. influenzae, bacilli disappeared when lysis buffer was added in all 3 BC systems (Figure 4). The mean score values of MALDI-TOF MS with or without lysis buffer were none and 1.921, respectively (Figure 5). By contrast, bacilli could be detected from all 3 BC bottles when we analyzed samples without lysis buffer, although RBCs were mixed (Table 1).
Detection of B. fragilis was similar to that of H. influenzae (Table 1).
Discussion
MALDI-TOF MS has revolutionized the identification of bacteria. It is fast, reliable, and inexpensive, and the majority of clinically relevant bacteria can be identified at the species level.3–5
To avoid any delay and misdiagnosis in bacterial identification during MALDI-TOF MS analysis, specialized software such as BioTyper (Bruker Daltonics) can be used, which has been shown to permit high-quality microbial identification, and some methods including RBC lysis have been performed.7
Here, we demonstrated the effects of adding lysis buffer in combination with several BC systems. BC broths from BD, bioMérieux, and Oxoid were prepared, and bacterial detection rates and MALDI-TOF MS scores were similar with and without lysis buffer for representative bacteria, such as E. coli, S. pneumoniae, and H. influenzae.
Furthermore, for E. coli, addition of lysis buffer led to clearer detection of E. coli (ie, increased MALDI-TOF MS scores) compared with analysis without the RBC lysis step. This finding may be because lysis buffer inhibits the effects of RBC mixing. In fact, the composition of BC broth, incubation atmosphere, and bacterial extraction method play key roles in the quality of further direct identification,5,8 and an in-house procedure for bacterial separation from BC broth using saponin has been developed.9 Similar results were observed for K. pneumoniae, and more effective detection of P. aeruginosa was found in our study when we used lysis buffer in MALDI-TOF MS analysis directly from BC.
However, S. pneumoniae was not detected by MALDI-TOF MS using the BD BC system when lysis buffer was added, and we could not detect any bacilli in this BC broth. However, regardless of the extraction method used in MALDI-TOF MS analysis, Gram-positive cocci are generally more difficult to detect than Gram-negative rods. Lysis buffer may destroy not only RBCs but also Gram-positive cocci, although Gram-positive cocci contain capsular polysaccharides and peptidoglycans. Moreover, Gram-positive cocci may be more permeable than Gram-negative rods that contain lipopolysaccharides and outer membranes. In addition, we could not detect S. pneumoniae without lysis buffer, suggesting that the lysis protocol may need to be optimized when using the BD BC system. Furthermore, S. pneumoniae were not identified without lysis buffer in the bioMérieux BC system, indicating the necessity of the lysis step in S. pneumoniae detection, similar to P. aeruginosa.
H. influenzae and B. fragilis were not detected in all 3 BC systems when lysis buffer was added, although we used Oxoid broth, which was the only system that could detect both S. pneumoniae and P. aeruginosa. Therefore, it may be better to use the pellet without resuspension as MALDI-TOF MS samples to avoid the risk of lysis buffer and RBC effects.
These differential effects of BC systems were also reported in the detection of fungal infection, including candidemia, by MALDI-TOF MS.10 The authors reported that direct BC on solid medium using a lysis–centrifugation procedure enabled successful Candida species identification by MALDI-TOF MS within an average of 3.8 hours (Sabouraud agar) or 7.4 hours (chocolate agar) before the positivity signal for control samples in BD BACTEC Mycosis-IC/F or BACTEC Plus Aerobic/F bottles, respectively. In addition, differential effects of lysis buffers were reported.11 The authors demonstrated that 2 inexpensive in-house detergent lysis methods, such as saponin and sodium dodecyl sulfate, were noninferior to a commercial kit, such as Sepsityper lysis buffer, for analysis of positive BCs by direct MALDI-TOF in a clinical diagnostic microbiology laboratory. The kinds of lysis buffer may not improve the ability of MALDI-TOF MS analysis of pathogens directly from BC broth.
Specificity/sensitivity of TOF-MS has been still an issue. It has been suggested that significantly more Gram-negative organisms were identified compared to Gram-positive organisms at species level.12,13 For example, monomicrobial BCs with Streptococcus oralis/mitis that were misidentified as S. pneumoniae, similar to our study.6 However, TOF-MS could detect the pathogens immediately at least at the genus levels, and finally contribute to early administration of antibiotics, survival of the patients, and cost-effectiveness. Therefore, we should use TOF-MS with caution, especially in sepsis with community-acquired pneumonia cases because S. pneumoniae and H. influenzae were representative pathogens of community-acquired pneumonia. Conventional methods, including BC and Gram staining, will still be mandatory.
In conclusion, we examined the effects of adding lysis buffer in the MALDI-TOF MS method to directly detect bacteria from 3 BC systems and compared detection efficiencies for specific pathogens. Detection of some bacteria in particular BC broths may be improved by adding a lysis step, especially for the Gram-negative pathogens, including E. coli, K. pneumoniae, and P. aeruginosa. However, caution may be needed for the detection process of S. pneumoniae and H. influenzae.
Protocols for individual species should be optimized to improve pathogen identification by MALDI-TOF MS analysis.
Acknowledgment
This work was supported by a Grant-in-Aid for Scientific Research (17K09623 to MS) from the Japanese Society for the Promotion of Science. (CC-BY-NC license was selected.)
Disclosure
The authors report no conflicts of interest in this work.
References
Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016;315(8):801–810. | ||
Yanagihara K, Kitagawa Y, Tomonaga M, et al. Evaluation of pathogen detection from clinical samples by real-time polymerase chain reaction using a sepsis pathogen DNA detection kit. Crit Care. 2010;14(4): R159. | ||
Moussaoui W, Jaulhac B, Hoffmann AM, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry identifies 90% of bacteria directly from blood culture vials. Clin Microbiol Infect. 2010;16(11):1631–1638. | ||
La Scola B, Raoult D. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One. 2009;4(11):e8041. | ||
Meex C, Neuville F, Descy J, et al. Direct identification of bacteria from BacT/ALERT anaerobic positive blood cultures by MALDI-TOF MS: MALDI Sepsityper kit versus an in-house saponin method for bacterial extraction. J Med Microbiol. 2012;61(Pt 11):1511–1516. | ||
Hariu M, Watanabe Y, Oikawa N, Seki M. Usefulness of matrix-assisted laser desorption ionization time-of-flight mass spectrometry to identify pathogens, including polymicrobial samples, directly from blood culture broths. Infect Drug Resist. 2017;10:115–120. | ||
Mellmann A, Bimet F, Bizet C, et al. High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J Clin Microbiol. 2009;47(11):3732–3734. | ||
Szabados F, Tix H, Anders A, Kaase M, Gatermann SG, Geis G. Evaluation of species-specific score cutoff values of routinely isolated clinically relevant bacteria using a direct smear preparation for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry-based bacterial identification. Eur J Clin Microbiol Infect Dis. 2012;31(6):1109–1119. | ||
Ferroni A, Suarez S, Beretti JL, et al. Real-time identification of bacteria and Candida species in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010;48(5):1542–1548. | ||
Idelevich EA, Grünastel B, Becker K. Rapid detection and identification of candidemia by direct blood culturing on solid medium by use of lysis-centrifugation method combined with matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). J Clin Microbiol. 2017;55(1):97–100. | ||
Tanner H, Evans JT, Gossain S, Hussain A. Evaluation of three sample preparation methods for the direct identification of bacteria in positive blood cultures by MALDI-TOF. BMC Res Notes. 2017; 10(1):48. | ||
Kok J, Thomas LC, Olma T, Chen SC, Iredell JR. Identification of bacteria in blood culture broths using matrix-assisted laser desorption-ionization Sepsityper™ and time of flight mass spectrometry. PLoS One. 2011;6(8):e23285. | ||
Lagacé-Wiens PR, Adam HJ, Karlowsky JA, et al. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J Clin Microbiol. 2012;50(10):3324–3328. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.