Back to Journals » OncoTargets and Therapy » Volume 11
Eukaryotic translation initiation factor 3 subunit G (EI F3G) resensitized HC T116/5-Fu to 5-fluorouracil (5-Fu) via inhibition of MRP and MDR1
Authors Yang C, Liu X, Li C, Li S, Du W, Yang D
Received 12 April 2018
Accepted for publication 12 June 2018
Published 31 August 2018 Volume 2018:11 Pages 5315—5324
DOI https://doi.org/10.2147/OTT.S170854
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Yao Dai
Chenggang Yang,1 Xin Liu,1 Chaobin Li,2 Shuangjing Li,3 Wenfeng Du,1 Daogui Yang1
1Department of Gastrointestinal Surgery, 2Department of Gastroenterology, 3Department of Central Laboratory, Liaocheng People’s Hospital, Liaocheng 252000, Shandong, China
Purpose: Colorectal cancer (CRC) has become a predominant cancer and accounts for approximately 10% of cancer-related mortality. Drug resistance still remains a priority mortality factor for patients due to no available therapeutic alternatives. The purpose of the present study was to investigate the underlying molecular mechanisms how eukaryotic translation initiation factor 3 subunit G (EIF3G) resensitized 5-Fu-resistant human CRC cells (HCT116/5-Fu) to 5-fluorouracil (5-Fu).
Methods: Multiple cellular and molecular biology experiments were performed in the present study, such as CCK-8, western blotting and flow cytometry.
Results: We found that EIF3G is highly expressed at RNA and protein levels in HCT116/5-Fu cells compared with HCT116 cells using quantitative real-time polymerase chain reaction and Western blot analysis. In addition, silencing EIF3G enhanced 5-Fu-induced apoptosis in HCT116/5-Fu cells. Moreover, EIF3G silencing decreased the activity of the drug-related proteins MDR1 and MRP levels in HCT116/5-Fu cells. Finally, the xenograft tumor model further confirmed that EIF3G resensitized HCT116/5-Fu tumors to 5-Fu. We observed that EIF3G silencing followed by 5-Fu administration had a synergistic interaction effect on HCT116/5-Fu in vitro and in vivo.
Conclusion: These findings demonstrate that EIF3G is a targetable regulator of chemoresistance in CRC, and inhibiting EIF3G in combination with 5-Fu might be a potential therapeutic strategy for colon cancer.
Keywords: EIF3G, colorectal cancer, 5-fluorouracil, drug resistance, MDR1, MRP
Introduction
Colorectal cancer (CRC) is ranked the third most common type of cancer and the fourth leading cause of cancer-related mortality in the world, with more than one million new cases diagnosed each year.1 Its etiology is multifactorial, and approximately 33% of all cases are attributed to genetic factors.2 In recent years, the increasing number of CRC in developed countries can take into account the increasingly aging population, unfavorable modern dietary habits, and other risk factors, such as smoking, low physical exercise, and obesity.3 At present, patients with stage I or II CRC are treated or cured by surgery. In patients with stage III CRC, the primary choice is combination treatment of surgical resection of tumor with chemotherapeutic agent or radiotherapy. 5-Fluorouracil (5-Fu) is one of the primary chemotherapeutic agents in clinical use.4 Despite advances in CRC research and treatment, CRC still remains incurable because of drug resistance both innate and acquired.
Eukaryotic translation initiation factors (EIFs) are the key factors that regulate translation initiation of protein synthesis in mammalian cells. EIF3 mediates the interaction between the 43S preinitiation complex and the eIF4F-bound mRNA clients.5 Eukaryotic translation initiation factor 3 subunit G (EIF3G) is one of the subunits of EIF3, which was reported to be involved in the process of translation reinitiation. Previous studies reported that EIF3G was involved in the caspase-mediated apoptosis6 and adriamycin-resistant human erythroleukemia cells.7 In addition, a positive correlation was observed between the overexpression of EIF3G and lymph node metastasis in breast cancer.8 Meanwhile, EIF3G has positive relationship with drug resistance-related proteins in Papaya ringspot virus.9
Many of the human ATP-binding cassette (ABC) proteins, such as MRP and MDR1 [also name P-glycoprotein (P-gp)], have been implicated to be the major efflux transporters responsible for multidrug resistance.10 MDR1 was the first investigated membrane transporter responsible to confer multidrug resistance to cancer cells. P-gp is encoded by the MDR1 gene following chemotherapy.11 MRP was discovered on the basis of non-P-gp-associated multidrug resistance.12 MDR1 and MRP have been widely investigated as multidrug resistance proteins and related to cancer resistance recently.
Considering the role of EIF3G involved in various stress response by enhancing the translation of resistance-related proteins, we hypothesized that the inhibition of EIF3G represents a novel strategy for overcoming drug resistance in CRC. In the present study, we demonstrate that silencing of EIF3G enhanced 5-Fu-induced HCT116/5-Fu cell apoptosis. More importantly, we discovered a role for EIF3G as a regulator for signal drug-related proteins MDR1 and MRP in vivo and in vitro. Our results indicated the influential role played by EIF3G in overcoming drug resistance in CRC.
Materials and methods
Cell culture
Human colon cancer HCT116 cell line was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). 5-Fu was purchased from Sigma Chemicals (St Louis, MO, USA). 5-Fu-resistant HCT116 (HCT116/5-Fu) was induced and established in our laboratory. In brief, HCT116/5-Fu cell line was established by culturing parental HCT116 cells with increasing concentration of 5-Fu (range from 0 to 50 μM). When the cells could survive at any given concentration of 5-Fu, they were passaged in drug concentrations that were 1.5- to 2-fold higher. Cells were grown in McCoy’s 5A medium supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), nonessential amino acids, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, Thermo Fisher Scientific) at 37°C and 5% CO2. All other reagents and solvents used in the following experiments were of analytical grade.
Cell proliferation assay
Cell proliferation assay was performed with the Cell Counting Kit-8 (CCK-8 assay kit, #96992, Sigma-Aldrich) per the manufacturer’s protocol. Briefly, HCT116 and HCT116/5-Fu (3×103 cells/well) were plated onto 96-well plate (100 μL medium of cell suspension) and then treated with various concentrations of 5-Fu with or without EIF3G silencing. Next, 10 μL of CCK-8 solution was added into each well of the plate after 24 hours. Finally, the cells were incubated for 30 minutes at 37°C, and the absorbance was measured at 450 nm using a microplate reader.
Quantitative real-time PCR
Total RNA was extracted from HCT116 cells and HCT116/5-Fu cells using (Trizol Invitrogen, Thermo Fisher Scientific) per the manufacturer’s protocol. Briefly, cDNA was carried out using 2 μg total RNA per sample using SuperScript II RNase H Reverse Transcriptase Kit (Life Technologies). Real-time polymerase chain reaction (PCR) was performed by using SYBR Green PCR mix (Thermo Fisher Scientific) with the following EIF3G primers: 5′-CTGGAGGAGGGCAAATACCT-3′ (sense) and 5′-CTCGGTGGAAGGACAAACTC-3′ (antisense). As control, the following GAPDH primers were used: 5′-ACACTCACTCTTCTACCTTC-3′ (sense) and 5′-TTGCTGTAGCCAAATTCATT-3′ (antisense). The quantitative real-time PCR procedure was performed in a Bio-Rad CFX Real-time System (Bio-Rad, Hercules, CA, USA) as follows: initial denaturation at 95°C for 10 minutes; 40 cycles of denaturation at 95°C for 15 seconds, and annealing extension at 60°C for 30 seconds. The relative expression of EIF3G was determined by the 2−ΔΔCt method, where Ct refers to cycle threshold. All samples were run in duplicate; mRNA expression level of EIF3G was normalized to the level of GAPDH.
Western blot analysis
Protein lysates were prepared by lysing the HCT116/5-Fu cell lines or tumor tissues in lysis buffer [50 mM Tris–HCl pH 6.8, 5 mM EDTA, 2% sodium dodecyl sulfate (SDS), and 5% glycine]. Protein concentration was estimated by BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts (30 μg) of protein samples were fractionated by SDS-polyacrylamide gel electrophoresis on a 12% polyacrylamide SDS gel at 80 V for 2 hours. Then, the proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) at 200 mA for 3 hours. The membranes were blocked for 1 hour with 5% skim milk in 0.1% TBST (20 mM Tris pH 7.6, 150 mM NaCl, and 0.05% Tween-20) followed by incubation with primary antibodies at 4°C overnight. The next day, the membranes were washed three times for 15 minutes in TBST, followed by incubation at room temperature with horseradish peroxidase-conjugated antirabbit or antimouse secondary antibody (Santa Cruz Biotechnology Inc., Dallas, CA, USA, dilution 1:5,000). Primary antibodies used rabbit antihuman EIF3G (Bethyl Laboratories, Inc., Montgomery, TX, USA; cat no A301-757A; 1:1,000), mouse antihuman GADPH (Santa Cruz Biotechnology; cat no sc47724; 1:1,000), mouse anti-MRP (Santa Cruz Biotechnology; cat no sc-136447; 1:1,000), and rabbit anti-MDR1 (Abcam, Cambridge, UK; cat no ab129450; 1:1,000).
Animal study
Six- to eight-week-old athymic nu/nu nude mice were obtained from Vital River (Beijing, China) and maintained under specific pathogen-free conditions. All experiments were performed under the approval of the Institutional Review Board at Liaocheng People’s Hospital (Liaocheng, China). The National Institutes of Health guide for the care and use of laboratory animals was strictly followed. The mice were randomly divided into two groups, and 5×106 HCT116/5-Fu cells transfected with EIF3G shRNA1 or control shRNA were subcutaneously injected. And the vehicle was administered to control mice; 30 mg 5-Fu was administered to both groups. Tumor volume was measured and calculated according to the formula (Tumor volume=length×width2×0.5).13 When the tumor volume reach about 180 mm3, the mice were intraperitoneally injected with 30 mg/kg 5-Fu once a week. The mice were sacrificed under deep isoflurane anesthesia after 4 weeks’ treatment, and the tumors were collected, photographed, and weighted. The body weight of tumor-bearing mice was monitored weekly.
Apoptosis assay
Apoptosis analysis was performed by Annexin V-FITC and propidium iodide (PI) staining using the Dead Cell Apoptosis Kit (Invitrogen, Carlsbad, CA, USA) per the manufacturer’s protocol. Briefly, treated cells were harvested by trypsin and washed twice in ice-cold PBS. Then, cells were fixed in 70% cold ethanol, centrifuged, washed with ice-cold PBS, and then resuspended in Annexin V/PI solution for 30 minutes at room temperature. Cells were analyzed using a flow cytometry (BD Biosciences, San Jose, CA, USA). Levels of apoptosis were calculated as the sum of early apoptotic cells (Annexin V positive/PI negative) and late apoptotic cells (Annexin V positive/PI positive).
EIF3G and MDR1 silencing
The following three EIF3G shRNAs were designed on Invitrogen Block-iT RNAi Designer (http://rnaidesigner.thermofisher.com/rnaiexpress/): EIF3G shRNA1, 5′-GCGGAATCGAATGAGATTTGC-3′; EIF3G shRNA2, 5′-GGAACATGTTGCAGTTCAACC-3′; and EIF3G shRNA3, 5′-GCCCTAGAATACTACGACAAA-3′. Nontarget shRNA 5′-GCGGAGGGTTTGAAAGAATTA-3′ was used as a negative control. Lentivirus particles were produced by transfecting HCT116/5-Fu cells with polybrene using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific). The stable negative control and EIF3G transfectants were selected and cultured in fresh medium supplemented with puromycin at 16 hours post-transduction. Forty-eight hours post-transduction cells were used for further experiments. The EIF3G silencing efficiency was analyzed by quantitative real-time PCR analysis. MDR1 siRNA (Thermo Fisher Scientific) was used to knockdown MDR1 expression in HCT116/5-Fu cell according to the manufacturer’s protocol.
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) Assay
TUNEL is a method widely used for detecting apoptotic cells.14 The tumor section slides were dewaxing, rehydration and inactivation endogenous peroxidase; the sections were incubated using TUNEL reaction mixture (In Situ Cell Death Detection Kit; Roche, Berlin, Germany) at 37°C for 60 minutes. TUNEL-positive cells were determined by two independent persons counting positive cells per visual field (5 fields per slide).
Statistical analysis
All experiments in the current study were done in triple. The SPSS 11.0 software (SPSS Inc., Chicago, IL, USA) was used to perform the statistical analysis. One-way analysis of variance was performed to determine the significance of differences between treatment groups. P<0.05 was considered statistically significant.
Results
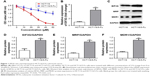
EIF3G was overexpressed in the HCT116/5-Fu cells
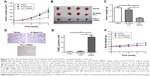
First, a 5-Fu drug-resistant HCT116 cell line had been established as described in Materials and methods, and their resistance to 5-Fu was confirmed by incubating with different concentrations of 5-Fu ranging from 0 to 50 μM (Figure 1A). Because EIF3G was involved in chemoresistance in some human cancer,7 we assessed its expression level in HCT116/5-Fu colorectal cells. Quantitative real-time PCR and Western blot analysis demonstrated EIF3G mRNA level, and protein expression was upregulated in HCT116/5-Fu cells than that in HCT116 cells (Figure 1B–D). Meanwhile, the protein levels of MRP and MDR1 were upregulated in HCT116/5-Fu cells compared with parental cells (Figure 1C–F).
EIF3G silencing enhanced 5-Fu-induced apoptosis in HCT116/5-Fu cells
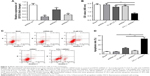
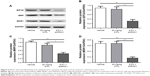
The short hairpin RNA was used to knock down EIF3G expression in HCT116/5-Fu cells. As shown in Figure 2A, EIF3G level was efficiently silenced by shRNA1 or shRNA3 in HCT116/5-Fu cells detected with quantitative real-time PCR. ShRNA1 exerted a better effect on silencing EIF3G than shRNA3 (Figure 2A). Thereby, EIF3G shRNA1 particles targeting EIF3G was used for the following experiments.
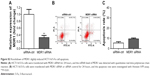
CCK-8 assay kit demonstrated that EIF3G silencing combined with 5-Fu significantly inhibited HCT116/5-Fu cell growth (Figures 2B and S1A). However, silencing EIF3G alone had very limited effects on HCT116/5-Fu cell viability (Figures 2B and S1B). To analyze whether the reduction in cell viability by 5-Fu plus EIF3G knockdown was related to apoptosis, the Annexin V/PI apoptosis analysis was performed. The data showed that EIF3G silencing enhanced 5-Fu-induced apoptosis in HCT116/5-Fu cells (Figure 2C and D). These results demonstrated that EIF3G silencing resensitized HCT116/5-Fu cells to 5-Fu.
Drug resistance-related proteins were downregulated by EIF3G silencing in HCT116/5-Fu cells
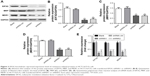
We next assessed the expression levels of drug resistance-related proteins (MRP, MDR1) with or without EIF3G silencing. The overexpression of P-gp encoded by MDR1 gene following chemotherapy severely limited the efficacy of anticancer agents.15 The Western blot results showed that MRP and MDR1 proteins were overexpressed in HCT116/5-Fu cells (Figure 1C), while silencing EIF3G using shRNA1 or shRNA3 significantly downregulated their levels (Figure 3A–D). Additionally, the mRNA levels of EIF3G, MRP, and MDR1 were also downregulated by shRNA1 or shRNA3 in the cells (Figure 3E).
As elucidated above, EIF3G silencing resensitized HCT116/5-Fu cells to 5-Fu via inducing apoptosis. Furthermore, the underlying mechanism of overcoming drug resistance was detected. The Western blot and quantitative real-time PCR results indicated that the protein and mRNA levels of MRP and MDR1 were not affected by 5-Fu alone treatment, while significantly decreased by EIF3G shRNA1 plus 5-Fu treatment (Figure 4A–E). In addition, knockdown MDR1 slightly induced HCT116/5-Fu apoptosis (Figure S2A–C).
EIF3G silencing resensitized HCT116/5-Fu tumor to 5-Fu in vivo
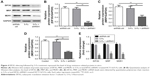
We next confirmed the role of EIF3G in vivo using HCT116/5-Fu xenograft mice model. In this model, 5-Fu alone treatment had a very limited effect on tumor growth, while the combination of 5-Fu with EIF3G silencing exerted a dramatically tumor growth inhibition (Figure 5A and B). Meanwhile, the quantification of tumor weight further confirmed the antitumor effect of EIF3G silencing plus 5-Fu administration (Figure 5C). Moreover, TUNEL assay was performed to evaluate the extent of apoptosis in tumor tissues. A significant increase in apoptotic positive cells was observed in the EIF3G silencing combined with 5-Fu treatment (Figure 5D and E). The body weight data of mouse indicated that 5-Fu or/and EIF3G shRNA1 treatments had very limited toxicity to mice (Figure 5F).
The Western blot analysis showed that MDR1 and MRP expressions were decreased by EIF3G silencing combined with 5-Fu treatment in tumor tissues. However, the expression levels of MDR1 and MRP had no change in 5-Fu treatment alone group (Figure 6A–D). These data were consistent with the results in vitro, suggesting the resensitization of HCT116/5-Fu model to 5-Fu through EIF3G silencing. In brief, these results demonstrated that EIF3G silencing and 5-Fu worked synergistically in 5-Fu-resistant xenograft in vivo.
Discussion
CRC is one of the most commonly diagnosed cancer worldwide.4 5-Fu was used as a single agent in patients who have an objective response rate of approximately 20%. The resistance to 5-Fu still remains the main problem in patients with CRC. Hence, to overcome the therapeutic resistance, the HCT116 cells resistant to 5-Fu were developed in the study. In the present study, we focused on investigating the role of EIF3G during CRC chemotherapeutic resistance development in vitro and in vivo.
Recently, EIF3G has been investigated as an oncogenic role in cancer. Previous studies have reported that EIF3G is involved in caspase-mediated apoptosis in T24 cells.6 Moreover, EIF3G was overexpressed in an adriamycin-resistant human erythroleukemia cell line.7 To our knowledge, this is the first time to elucidate the relationship of EIF3G with drug resistance in CRC. In the current study, EIF3G was markedly upregulated in HCT116/5-Fu cells compared with HCT116 cells (Figure 2). Silencing EIF3G followed by 5-Fu treatment induced apoptosis and resensitized to HCT116/5-Fu to 5-Fu. This was further corroborated by in vivo mice xenograft study, which showed that tumor volume and tumor weight of animal were dramatically inhibited by EIF3G silencing plus 5-Fu administration. These data provided clues to further investigate the molecular mechanisms underlying the multidrug resistance properties of EIF3G silencing in HCT116/5-Fu cells.
In recent years, drug efflux pumps, MDR1, and MRP have been widely investigated in association with chemotherapeutic resistance in CRC.16,17 In the present study, we found that EIF3G silencing significantly decreased the levels of MDR1 and MRP (Figure 3) in HCT116/5-Fu. Similar results were observed in EIF3G silencing followed by 5-Fu treatment groups in vivo. However, S´roda-Pomianek et al reported that the level of MDR1 was reduced to overcome resistance by oxicam derivative in CRC.17 Thus, we hypothesize that EIF3G silencing resensitized HCT116/5-Fu cells to 5-Fu via regulating the efflux transporters of MDR1 and MRP drug efflux. Meanwhile, it was also possible that EIF3G regulated MRP and MDR1 expression in a transcriptional level because the mRNA levels of MRP and MDR1 were downregulated by EIF3G shRNA1. However, the detailed mechanisms were still unclear and needed further investigation in future.
Conclusion
Our findings indicated that EIF3G was upregulated in 5-Fu-resistant HCT116 CRC cells. EIF3G silencing followed by 5-Fu treatment resensitized HCT116/5-Fu cells to 5-Fu via inducing apoptosis in vitro and in vivo. Our findings demonstrate that EIF3G is a targetable regulator of chemoresistance in CRC, and hence inhibiting EIF3G in combination with 5-Fu is a potential approach for colon cancer.
Acknowledgment
This study was supported by Shandong Provincial Natural Science Foundation (China ZR2015HL082).
Disclosure
The authors report no conflicts of interest in this work.
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. | ||
Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. | ||
Kuipers EJ, Grady WM, Lieberman D. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. | ||
Papamichael D, Audisio RA, Glimelius B, et al. Treatment of colorectal cancer in older patients: International Society of Geriatric Oncology (SIOG) consensus recommendations 2013. Ann Oncol. 2015;26(3):463–476. | ||
Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–266. | ||
Kim JT, Lee SJ, Kim BY, et al. Caspase-mediated cleavage and DNase activity of the translation initiation factor 3, subunit G (eIF3g). FEBS Lett. 2013;587(22):3668–3674. | ||
Zhu F, Wang Y, Zeng S, Fu X, Wang L, Cao J. Involvement of annexin A1 in multidrug resistance of K562/ADR cells identified by the proteomic study. OMICS. 2009;13(6):467–476. | ||
Zheng Q, Liu H, Ye J, Zhang H, Jia Z, Cao J. Nuclear distribution of eIF3g and its interacting nuclear proteins in breast cancer cells. Mol Med Rep. 2016;13(4):2973–2980. | ||
Gao L, Tuo D, Shen W, Yan P, Li X, Zhou P. NIa-Pro of Papaya ringspot virus interacts with Carica papaya eukaryotic translation initiation factor 3 subunit G (CpeIF3G). Virus Genes. 2015;50(1):97–103. | ||
Glavinas H, Krajcsi P, Cserepes J, Sarkadi B. The role of ABC transporters in drug resistance, metabolism and toxicity. Curr Drug Deliv. 2004;1(1):27–42. | ||
Roundhill E, Burchill S. Membrane expression of MRP-1, but not MRP-1 splicing or Pgp expression, predicts survival in patients with ESFT. Br J Cancer. 2013;109(1):195–206. | ||
Cole SP, Bhardwaj G, Gerlach JH, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258(5088):1650–1654. | ||
Hosaka K, Yang Y, Nakamura M, et al. Dual roles of endothelial FGF-2-FGFR1-PDGF-BB and perivascular FGF-2-FGFR2-PDGFRβ signaling pathways in tumor vascular remodeling. Cell Discov. 2018;4:3. | ||
Lozano GM, Bejarano I, Espino J, et al. Relationship between caspase activity and apoptotic markers in human sperm in response to hydrogen peroxide and progesterone. J Reprod Dev. 2009;55(6):615–621. | ||
Baker EK, Johnstone RW, Zalcberg JR, El-Osta A. Epigenetic changes to the MDR1 locus in response to chemotherapeutic drugs. Oncogene. 2005;24(54):8061–8075. | ||
Mudududdla R, Guru SK, Wani A, et al. 3-(Benzo[d][1,3]dioxol-5-ylamino)-N-(4-fluorophenyl)thiophene-2-carboxamide overcomes cancer chemoresistance via inhibition of angiogenesis and P-glycoprotein efflux pump activity. Org Biomol Chem. 2015;13(14):4296–4309. | ||
Środa-Pomianek K, Wesołowska O, Szczęśniak-Sięga B, et al. Effect of new oxicam derivatives on efflux pumps overexpressed in resistant a human colorectal adenocarcinoma cell line. Anticancer Res. 2015;35(5):2835–2840. |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.