Back to Archived Journals » Gastrointestinal Cancer: Targets and Therapy » Volume 4
Esophageal cancer-related gene 4 at the interface of injury, inflammation, infection, and malignancy
Authors Baird A, Lee J, Podvin S, Kurabi A, Dang X, Coimbra R, Costantini T, Bansal V, Eliceiri BP
Received 22 July 2014
Accepted for publication 28 August 2014
Published 24 October 2014 Volume 2014:4 Pages 131—142
DOI https://doi.org/10.2147/GICTT.S49085
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Andrew Baird, Jisook Lee, Sonia Podvin, Arwa Kurabi, Xitong Dang, Raul Coimbra, Todd Costantini, Vishal Bansal, Brian P Eliceiri
Division of Trauma, Burn, and Acute Critical Care, Department of Surgery, University of California San Diego Health Sciences, San Diego, CA, USA
Abstract: In humans, esophageal cancer-related gene 4 (ECRG4) is encoded by four exons in the c2orf40 locus of chromosome 2. Translation of ECRG4 messenger ribonucleic acid produces a 148 amino acid-secreted 17 KDa protein that is then processed to 14, ten, eight, six, four, and two KDa peptides, depending on the cell in which the gene is expressed. As hypermethylation at the c2orf40 locus inhibits ECRG4 gene expression in many epithelial cancers, several investigators have speculated that ECRG4 is a candidate tumor suppressor. Indeed, overexpression of ECRG4 inhibits cell proliferation in vitro, but it also has a wide range of effects in vivo beyond its antitumor activity. ECRG4 overexpression affects apoptosis, senescence, cell migration, inflammation, injury, and infection responsiveness. ECRG4 activities also depend on its cellular localization, secretion, and post-translational processing. These cytokine/chemokine-like characteristics argue that ECRG4 is not a traditional candidate tumor suppressor gene, as originally predicted by its downregulation in cancer. We review how insights into the regulation of ECRG4 gene expression, knowledge of its primary structure, and the study of its emerging physiological functions come together to support a much more complex role for ECRG4 at the interface of inflammation, infection, and malignancy.
Keywords: ECRG4, c2orf40, tumor suppressor, cytokine, chemokine, growth inhibitor
Introduction
An estimated 25,000 human genes are being assigned to distinct loci in human chromosomes, and often, their functions are not known. Of these genes, open reading frames (ORFs) are encoded by only a small fraction of genes that, in turn, transcribe messenger ribonucleic acid (mRNA) to produce proteins. An even smaller subset of ORFs encode extracellular proteins. These are designated the human “secretome”,1,2 and they encode peptide ligands, cell surface proteins, high- and low-affinity receptors, and circulating enzymes that naturally localize outside the cell. Because of this pharmacological accessibility, these gene products are attractive targets for diagnostics development and new therapeutics discovery.
In a search of genes encoding peptide hormones in the human genome, Mirabeau et al3 used Markov modeling to identify one likely candidate, the human c2orf40 locus of chromosome 2, ORF 40 (c2orf40). Interestingly, this same locus was previously reported4,5 to encode a candidate tumor suppressor gene called esophageal cancer-related gene 4 (ECRG4). Nevertheless, Mirabeau et al3 discovered that the ECRG4 gene encoded a protein with all of the features of a secreted neuropeptide hormone precursor. At the time, ECRG4 had no known physiological function, but the ECRG4 protein was recognized to have a hydrophobic leader sequence targeting it to the endoplasmic reticulum and Golgi apparatus, and thereby its secretion outside the cell. At 148 amino acids in length, Mirabeau et al3 observed that ECRG4 also had a primary sequence that was highly conserved among species. Finally, with a molecular weight of 17,183 Daltons, ECRG4 was relatively small, and its primary amino acid sequence contained consensus dibasic amino acids that predicted the same proteolytic post-translational processing that characterized many extracellular ligands.6–9
Because ECRG4 expression is significantly downregulated by hypermethylation in human cancers,4,5,10–22 there are compelling reasons to believe that its basic biology might be directly tied to the onset, development, and progression of human cancer. If so, the physiological methylation of its promoter in normal cells may gauge its expression in normal tissues and fine-tune ECRG4 gene re-expression after injury, inflammation, and infection, and its effects on apoptosis, cell migration, and senescence.3,23–39
Discovery of esophageal cancer-related gene 4
In 1998, Su et al4 described how cloning and sequencing expressed ribonucleic acids could be used to implicate genes in the development of esophageal cancer. Using a technique of differential display, they compared differences in gene expression between three normal and three primary human squamous esophageal cell carcinomas, and identified four novel genes that were either expressed in normal esophageal epithelia but absent in esophageal cancer or alternatively expressed in esophageal cancer but not detected in normal esophageal epithelia. These esophageal cancer-related genes were named ECRG1, ECRG2, ECRG3, and ECRG4, and further studied by reverse transcription polymerase chain reaction. All were found in many tissues, including the fetal and adult brain, liver, kidney, testis, bone marrow, and skeletal muscle. ECRG1 and ECRG2, however, were not detectable in either cancerous or cancer-adjacent tissues, while ECRG3 was highly expressed. ECRG4 gene expression appeared unique in that it was decreased in tumor cells, but readily detectable in normal tumor-adjacent tissue. Su et al4 concluded that ECRG1 and ECRG2 were most likely tumor associated,4 so the role of ECRG4 was largely ignored.
A subsequent bioinformatic approach by Bi et al,5 however, supported a broader role for ECRG4 in the control of cancer cell growth. They confirmed that ECRG4 was downregulated and extended their findings to human prostate cancer and several tumor cell lines. They also mapped the ECRG4 gene to human chromosome 2q14.1–4.3, a finding that would later be corrected to 12q12.2 and defined to the c2orf40 locus.
The field remained largely dormant until 2002, when investigators detected differential ECRG4 gene expression in the course of screening candidate genes implicated in epithelial dysfunction and inflammatory disease.23–25 These were the first indications that ECRG4 might be involved in homeostatic functions in normal physiology. These authors also confirmed that ECRG4 gene expression was readily detectable in numerous tissues, including human heart, brain, placenta, lung, liver, skeletal muscle, eye, cornea, and kidney. While its physiological function remained enigmatic, at least one report implicated ECRG4 in tissue regeneration after injury,26 a finding that would be confirmed several years later in preclinical models of central nervous system (CNS) injury, infection, and immunity.30,35,37–40
The potential biological relevance of ECRG4 in glaucoma41 extended the interest of ECRG4 in malignancy and suggested that ECRG4 had a broader regulatory function in human biology. It was the work of Su et al,10 however, in 2003 that confirmed the potential importance of ECRG4 in human cancer biology. These authors reported that ECRG4 gene expression profiles significantly correlate (P=0.001) with family histories of upper gastrointestinal cancers.10 Their study was followed shortly thereafter by the discovery that ECRG4 downregulation is mediated by CpG hypermethylation of its promoter.11 This hypermethylation, in turn, is a mechanism for regulating ECRG4 gene expression based on de-methylation drugs that restore its expression.37,42
By 2007, the detection of ECRG4 had gained value as an independent prognostic factor of poor survival in patients with esophageal cancer12 and implicated in tissue regeneration after injury.26 Just as Markov modeling predicted,3 ECRG4 was secreted. Peptides processed from the ECRG4 precursor were detected in proteomic analyses of normal human biological fluids like blood, cerebrospinal fluid2,31,34,43 and in the conditioned media of different cell types.44,45 Unlike “traditional” tumor suppressor genes, ECRG4 was also tethered to the outer cell surface and processed for release into biological fluids.37,44,45 Clearly, even though ECRG4 was most often described as a “candidate tumor suppressor gene”, it was better described as a neuropeptide, cytokine, and chemokine7,23 implicated, by virtue of its downregulation, in the onset and progression of human cancers.
The ECRG4 gene: expression, regulation, and hypermethylation
In humans, the ECRG4 protein is encoded by a four-exon gene on chromosome 2 at the locus C2:106,063,246 to 106,078,159 that is called C2orf40 (Figure 1). Bioinformatic mapping of the 5′ untranslated region (5′UTR) reveals a significant distribution of CpG islands that are predicted substrates for hypermethylation of the c2orf40 promoter. This structural finding is compatible with a very strong (P=0.001) correlation between the inhibition of ECRG4 gene expression and its hypermethylation in cancer.11,13–15,21 It is further supported by the fact that methylation inhibitors like the drug 5-AZA-C, can increase ECRG4 expression in vivo and in vitro,37,42 presumably by inhibiting deoxyribonucleic acid (DNA) methylation. Although there is a significant body of evidence pointing to methylation of ECRG4 promoter CpGs, their exact identity and the significance of individual methylation sites remains unknown. Bioinformatic promoter analyses of the 5′UTR of the c2orf40 gene also identify several consensus transcription factor binding sites that predict a role for numerous intracellular transcription factors in the regulation of ECRG4 expression. Interestingly, many of these transcription factors are associated with the control of cell growth and differentiation, suggesting the possible roles of ECRG4 in cell function (Figure 1). These include SP1, Ik-2, cRel, SRY, RORα2, Oct-1, MZF-1, c-myc, Ap1, and AML1α. As expected, a TATA transcription start sequence is localized 1 Kb 5′ from the ECRG4 open reading frame in the 5′UTR.
  | Figure 1 The ECRG4 gene and the ECRG4 promoter. |
As discussed earlier, the ECRG4 gene is normally expressed in a wide range of normal tissues that include adrenal, bone marrow, brain, breast, esophagus, eye, heart, intestine, kidney, leukocytes, lung, ovary, placenta, prostate, skeletal muscle, skin, spleen, testis, and thyroid. Because of this wide tissue distribution, it can be presumed to have constitutive functions37,39,40,46 maintaining homeostasis. These sentinel functions are only beginning to emerge and are discussed below. To date, ECRG4 gene expression has been primarily localized to epithelial cells, including more specialized epithelial-derived cells and even hematopoietic cells. These include pituicytes,47,48 oligodendrocytes,30 astrocytes,43 choroid plexus epithelium, and ventricular ependymal cells35,40 in the CNS, and mucosal,39 epidermal,38 corneal,24 and airway epithelial cells49 and chondrocytes25,28 outside the CNS. Remarkably, ECRG4 is also detected on the surface of circulating neutrophils and on lymphocytes37 derived from bone marrow4 and spleen.24
In contrast to the wide distribution of ECRG4 gene expression in normal tissues, the epigenetic silencing of ECRG4 expression by methylation of its promoter downregulates constitutive human ECRG4 gene expression in a wide variety of human cancers.11,13–15,18,21,37,40,42 In these patients, the correlation between ECRG4 expression and outcome appears to be particularly important. Higher ECRG4 expression levels (like those seen in normal tissues) correlates with improved survival in patients with cancers of the esophagus, prostate, and breast. These findings support a proposed role for ECRG4 as a candidate tumor suppressor12,15,19 in regards to cancer, but equally as a sentinel gene37,39,40,46 in normal tissues. To this end, it is important to note that ECRG4 expression confers increased cell sensitivity to antitumor drugs like 5-fluorouracil, presumably by virtue of its ability to increase apoptosis.20 Interestingly, there are also low to undetectable levels of ECRG4 gene expression in a wide panel of cultured normal cells or alternatively expanded as tumor cell lines. While we have attributed this change to the injury phenotype that alters ECRG4 gene expression (see “Infection, inflammation, and injury”), others28 attribute the observation of undetectable ECRG4 protein to the sensitivity of reagents. In part, this decreased gene expression may be due to promoter hypermethylation, since gene expression can be normalized by treating cells with DNA methylation inhibitors such as 5-Azacytosine.37
Sequence, secretion, and cell-specific processing
The primary peptide sequence of ECRG4 is highly conserved among vertebrates, suggesting that ECRG4 plays an important physiological function that is shared by different species. This is best illustrated by the high degree of homology between the ECRG4 of different species (Table 1) but is shown in Figure 2A as sequence conservation that can be traced to specific domains in the protein. This homology may thus infer possible physiological importance, point to mechanism of action, and help establish the protein’s relevance to disease. For example, the C-terminus of the ECRG4 “precursor” is strikingly conserved among all species, and interestingly, a putative thrombin consensus sequence in human ECRG4 (126–135) appears to be absent in lower species. This could suggest taxonomically restricted biological activities due to species-specific protein processing.
  | Table 1 Homology of ECRG4 in different species |
As discussed above, bioinformatic analyses also predict that the human ECRG4 open reading frame, ECRG4, encodes a secreted protein that has a hormone precursor-like structure that is susceptible to proteolytic processing.3 Processing can generate multiple ECRG4-derived peptides and, like many neuropeptide precursors6–8,50 each peptide can have individual, shared, and biologically distinct activities depending on proteolytic processing (Figure 2B). To this end, several studies have shown that ECRG4-derived peptides can be detected in lysates of normal tissues, in biological fluids (eg, serum and cerebrospinal fluid), and the conditioned media of cells engineered to overexpress ECRG4.13,28,30,31,37,40,43–45,51 Immunohistochemistry, flow cytometry, and subcellular localization studies, however, demonstrated that, while ECRG4 is indeed secreted outside the cell, in some cells it remains tethered to the cell surface.45 Cell surface biotinylation assays, mutagenesis analyses, and confocal microscopy show that its release from tethering is cell type-dependent (Figure 2B). Human kidney epithelial cells display the 14 KDa protein tethered to their surface and it is not further processed. Human neutrophils37 constitutively express cell surface ECRG4 and require cell activation to release the 14 KDa peptide that is processed to smaller peptides (Figure 2B). The PC3 prostate cancer cell line appears to display ECRG4 on the cell surface, but releases the peptide constitutively.45 A near-ubiquitous distribution of the membrane-bound 14 KDa ECRG4 in largely quiescent tissues in vivo, however, suggests that the precursor serves a sentinel-like function that might also be constitutively proquiescent and growth/migration inhibitory. Upon cell stimulation and cell surface activation by prohormone convertases, peptides produced by ECRG4 processing can therefore play synergistic, potentiating, or even opposing functions and be growth/migration inhibitory, anti-inflammatory, and/or proinflammatory, respectively. This may be why downregulated and upregulated ECRG4 is associated with both pro- and anti-inflammatory activities, and pro- and antiapoptotic responses that alternatively stimulate cell senescence, affect the stress response, and promote cell survival.13,28,30,31,37,40,43–45,51
  | Figure 2 ECRG4 protein, processing, and secretion by the cell. |
The processing and release of ECRG4 derived from the cell surface is presumed to be dependent on enzymatic processing at putative PC1, PC2, and PC3 furin-like cleavage sites in the primary sequence of ECRG4. However, the specific enzymes responsible for ECRG4 processing at the cell surface have yet to be identified. It is particularly interesting to note that the ECRG1 gene,4 which was originally linked to esophageal cancer at the same time as ECRG4, encodes a transmembrane serine protease that interacts with ECRG4.52 This ECRG1 gene, now called TMPRSS11A, is a proteolytic enzyme that, like ECRG4, is associated with epithelial cancer progression, cell senescence, the inhibition of cell growth, and G1 cell cycle arrest. These are all biological activities that it shares with ECRG4, suggesting that its downregulation in esophageal cancer4 would impair release and activation of an antitumor ECRG4 from the cell surface. While their interaction in normal tissues is unknown, it is interesting to speculate that ECRG1 might be a physiological activator of cell surface ECRG4 in vivo.
Inflammation and tumor suppression
The original discovery of ECRG4 downregulation in cancer by Su et al4 implied that it might be a constitutively expressed autocrine cell growth inhibitor. Indeed, several reports since its discovery have described in vitro inhibitory activities of ECRG4 gene overexpression,14,17,21,22,53 but to date there is still no report demonstrating growth inhibitory biological activity of recombinant ECRG4 proteins.54 In contrast, others have shown ECRG4 overexpression and ECRG4 peptides to be either stimulatory, proinflammatory, or to require proteolytic processing for activity.27,30,32,37,44 The discovery of ECRG4 expression in human leukocytes in particular37 raised the possibility that its association with tumor growth might include both epithelial and immuno-inflammatory signaling. Together, these findings support the observation that ECRG4 is unlike other traditional and more commonly studied tumor suppressor genes like p53 and Rb, although they likely interact.55 In addition to being secreted, encoding a ligand, and being processed differently in different cell types, there are also no reports of genetic mutations in the c2orf40 locus that are associated with human cancers. While some sequence variations have been detected in a small fraction of breast tumors, the loss of ECRG4 in cancer is most likely attributable to the incidence of decreased gene expression rather than inactivating polymorphisms.19 These observations are also compatible with those describing a downregulation of ECRG4 gene expression that is primarily epigenetic and therefore mediated by promoter hypermethylation.13–15
Although several reports describe ECRG4 with implied tumor suppressor activity, the mechanism of any of these antitumor activities remains largely unknown. For example, no specific peptide has unequivocally been shown to be responsible for the biological effects of ECRG4 gene overexpression. Accordingly, its activities remain contentious. Overexpression of ECRG4 in vivo has significant effects on cell growth and function in several preclinical models of human disease (see “Infection, inflammation, and injury”). Whereas some studies have suggested a direct antitumor cell effect in vitro, however,14,17,18,52 the results have not been convincing or confirmed and, in at least two instances, the published research papers54,56 have been retracted. In our laboratories, we have had significant difficulty establishing a direct antimitogenic effect of ECRG4 overexpression when transfecting tumor cells in vitro.57 Yet in vivo, tumor studies reproducibly show significant antitumorigenic effects of ECRG4 in orthotopic tumor models of CNS and subcutaneous cancers.57 We speculate that this variability is due to the need for proteolytic processing to observe biological activity,44 and that proteolytic processing is very cell specific.45 Other investigators have pointed to indirect mechanisms that might account for in vivo antitumor activities, for example by increased autocrine induction of apoptosis20 or the proinflammatory effects of a small 16-amino-acid peptide that is released from the C-terminus of ECRG4.37
These findings add credence to the observations that ECRG4 would have to be unique among “candidate” tumor suppressor genes because it is extracellular, localized to the cell surface, and shed into extracellular milieu. Tumor suppressor genes, in contrast, usually encode intracellular proteins that serve as transcription factors and components of intracellular signaling pathways. ECRG4 activities appear more reminiscent of cytokine-chemokine-growth factors in that they also require proteolytic processing for activity. These similarities would also suggest that ECRG4 acts via paracrine pathways, and most likely in conjunction with circulating immune and inflammatory cells37 through mechanisms more similar to immunosurveillance than tumor suppression. Leukocytes are a rich natural source of ECRG4, and a thrombin-processed, 16-amino-acid peptide is a chemoattractant of myeloid cells.57 Overexpression of ECRG4 in preclinical models of gliomas reverses the immunosuppressive tumor microenvironment, reduces the glioma burden, and increases in the survival of glioma-bearing animals.57 These observations are markedly similar to at least two other cytokine-like factors with similar ECRG4-like antitumor activity. Both thymic stromal lymphoprotein and galectin, for example, are outside the cell, localize to the cell surface, and influence myeloid cell infiltration into tissues.58–60 On the basis of these similarities, the findings with ECRG4 are consistent with the hypothesis that it is first a physiological immunomodulatory/immunosurveillance factor37 that, when introduced into the tumor bed, regulates the tumor immune microenvironment and the control of tumor growth.
Infection, inflammation, and injury
Just as ECRG4 gene expression is downregulated in tumors, it is also seen downregulated after injury and infection. In a model of mucosal infection, for example (Figure 3), ECRG4 is constitutively expressed, but gene expression decreases rapidly following infection at times that immediately precede cell proliferation. The return of ECRG4 gene expression also precedes the return to quiescence. Similar responses are observed in other models of acute injury,25,28,35,37,39,40 although increased ECRG4 gene expression is reported in some models of chronic inflammation,25,28 including transgenic mice that overexpress the tau23 protein to mimic Alzheimer’s disease.61 Shaterian et al38 also showed that there was a slow and graded elevation in ECRG4 gene expression in skin, possibly due to the invasion of ECRG4+ leukocytes37 that mobilize to the wound after excisional cutaneous injury, and gene expression was highest at the last stages of wound resolution. This finding is compatible with an ECRG4 participation in the regulation of epithelial to mesenchyme transition that occurs in wound healing,38 and by extension, perhaps, in cancer. To date, this mechanism has not been explored. Instead, in models of CNS injury, there is a rapid decrease in constitutive ECRG4 gene expression that decreases sharply by 24 hours, but is transient and normalizes shortly thereafter. The decrease in ECRG4 is coincident with a peak in neuroprogenitor cell proliferation and the ECRG4 response appears dependent on the severity of the CNS lesion, suggesting a physiological role for ECRG4 in gauging parenchymal and inflammatory response to traumatic brain injury. In these studies, a small needle injury leads to a transient decrease, with a full recovery of ECRG4 within 24–48 hours.40 In contrast, a larger penetrating injury35 results in more sustained decreases in ECRG4 expression to 72–120 hours, although a full recovery is observed shortly thereafter. Interestingly, a forced and continuous expression of ECRG4 using adenovirus encoding ECRG4 compensates for the injury-induced loss of ECRG4 gene expression and alters the natural course of the injury response.35,39,40 These experiments establish a causal relationship between the decrease in ECRG4 expression and injury-induced changes, like the increase in cell migration in vivo.35,40 They also infer that processes regulating endogenous gene expression will impact the response to injury. It is therefore interesting to note that these findings predict that the epigenetic methylation of the ECRG4 promoter will gauge ECRG4 gene responsiveness to injury by controlling basal gene expression and its ability to return to levels post injury.
  | Figure 3 Injury-induced downregulation of ECRG4. |
In a separate set of experiments, Kurabi et al39 studied mucosal epithelial cells because they are highly sensitive to inflammatory cytokines in vivo and in vitro, and respond to infection with reversible mucosal thickening and cell proliferation. Like other tissues, ECRG4 gene expression is abundant in normal mucosal epithelia, but it decreases after infection and inflammation.39 At resolution, ECRG4 gene expression returns to preinfection quiescent levels and epithelial thickening and proliferation normalize to baseline. Although the mechanisms that restore ECRG4 expression have not been investigated, the ECRG4 promoter has many characteristics of a “housekeeping” gene (Figure 1) that predict constitutive expression (eg, Sp1 consensus sites within CpG islands). No studies have been published regarding the possible transcription factors that transiently regulate gene expression after injury or infection as opposed to its epigenetic silencing by hypermethylation. Nevertheless, this regulation appears critical because the delivery of exogenous ECRG4 with adenovirus prevents the observed decrease in tissue ECRG4 and blocking the inhibition protects the epithelium from injury39 and establishes that ECRG4 plays a functional role in epithelial responsiveness to infection. Furthermore, these data support its candidacy as a sentinel gene that gauges inflammation through immunosurveillance rather than a traditional tumor suppressor gene.
The role of ECRG4 in normal development has also been explored, albeit to a limited extent, in the developing brain. There, ECRG4 is selectively upregulated in the choroid plexus and ependyma throughout development, and this expression is maintained in the adult.40 During aging, ECRG4 gene expression naturally increases, and because its overexpression in oligodendrocyte precursor cells induces senescence, it may play a physiological role in aging.30 Knockdown of ECRG4 gene expression during zebrafish development is accompanied by ventriculomegaly that has not been explained, but an overproliferation of astrocytes suggests that ECRG4 regulates cell growth in vivo.40 In bone, ECRG4 expression fluctuates with the differentiation state of the chondrocyte,28 and expression is very low in mesenchymal cells but dramatically increases during chondrogenic differentiation.28 In as much as they mirror an injury response, ECRG4 levels are also decreased in chondrocytes derived from human osteoarthritic cartilage and in tissues of experimental mouse osteoarthritic cartilage.28
Signal transduction
With two exceptions,32,37 the efforts to characterize the signal transduction pathways that are activated by ECRG4 have relied on transducing cells to overexpress the ECRG4 gene and then examine changes in cell signaling pathways. However, because the full identification of naturally processed ECRG4-derived peptides has remained elusive,3 it is impossible to determine which pathways are activated by different ECRG4 derived peptides. The synthetic peptide ECRG4133–148 and the recombinant protein ECRG473–148 appear proinflammatory,32,37 but ECRG4 gene overexpression shows both pro- and anti-inflammatory responses depending on the model used. There can be increased or decreased nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) expression, pro- or antiapoptosis, and an inhibition of cell growth or differentiation. Presumably these mixed results are attributable to the fact that different transfected cells produce different ECRG4-derived peptides with potentially different activities and signaling capacity.3,37,44
That being said, because ECRG4 gene expression is low in undifferentiated mesenchymal cells but dramatically increased during chondogenesis and then decreased in hypertrophy,28 there is a general agreement that gene expression is dynamically regulated during development as it is after injury. On one hand, Li et al14,17 reported ECRG4 transfection inhibits proinflammatory NF-κB activation, its nuclear translocation, and the activation of cyclooxygenase-2 as a mechanism to inhibit cell proliferation, colony formation, anchorage-independent cell growth, and cell cycle progression. Yet in the CNS, Kujuro et al30 reported increased senescence mediated by increased cyclin D1 and D3 degradation and signaling. These data are compatible with the regulation of ECRG4 by the tumor suppressor p5355 but are in contrast to its upregulation in the proinflammatory milieu like those caused by Tau23 overexpression.61 As protein processing appears required for inhibitory ECRG4 activities,44 it is difficult to ascribe and specifically differentiate the involvement of signal transduction pathways that implicate NF-κB or transcription pathways like Pou3f436 and ETS,62 Fas and caspase 8,63 or alternatively the ubiquitin conjugating enzyme UBE2C.21
Clinical significance of ECRG4 to human disease
The data that link the downregulation of ECRG4 gene expression to the development and progression of epithelial cancers are now significant.4–6,11–13,16–22,44,52,53,55,63,64 Furthermore, its normal homeostatic functions in fetal development and in injury, inflammation, and infection responses are emerging.24–30,33,35–41,43,45,65 Because ECRG4 is unlike other candidate tumor suppressor genes in that it is secreted, tethered to the surface, and proteolytically processed for biological activity, its biology offers significant and proven potential for new drug discovery, like inhibitors of promoter hypermethylation to upregulate gene expression, new drug-target discovery like agonists to its high affinity receptors, drug development like protease inhibitors of cell surface processing, and biomarker assessment like flow cytometric analyses of circulating cells. In previous studies,35,37–40 we suggested that although an ECRG4 tethered to the cell surface could act as an anti-inflammatory sentinel factor that gauges the injury response, it could have opposing proinflammatory activities when processed. In this model (Figure 4), ECRG4 is present on the cell surface of both quiescent epithelial cells35,38–40 and circulating leukocytes.37 This cell surface ECRG4 expressed in quiescent tissues (Figure 4A) may then have a “sentinel” function that monitors homeostasis, gauges the proinflammatory response to injury and infection,35,39,40 and thereby maintains quiescence. Upon infection or inflammation (Figure 4B), protease activation and priming releases processed forms of ECRG4 from the cell surface37,44,45 and ECRG4 gene expression is downregulated.35,39,40 This gives repair cells an ECRG4 cancer “genotype” and proliferative “phenotype.”4–6,11–14,16–22,52,53,55,63 The release of smaller processed ECRG4 peptides have intrinsic activities that recruit, inhibit, and activate both inflammatory and epithelial cells.37,44 These reactive epithelial cells (Figure 4C) are characterized by a downregulated ECRG4 gene expression genotype and a loss of ECRG4 on the cell surface. This facilitates responsiveness to autocrine, paracrine, and endocrine growth promoting factors (Figure 4D) that promote increased cell migration and proliferation.35,39,40 Blocking the loss of ECRG4 prevents this process.35,39,40 The re-expression of intact ECRG4 to the cell surface in a protease-free or protease-neutralized environment would be dependent on promoter activity and its methylation state. As ECRG4 expression returns, it would inhibit cell responsiveness (Figure 4C) and reactivity (Figure 4B) to help restore quiescence (Figure 4A). In cancer, the continuous downregulation of ECRG4 through hypermethylation blocks the restoration of ECRG4 to the cell surface and as such compromises normal immune cell surveillance (Figure 4A), resulting in the display of a continuous reactive phenotype (Figure 4B) that enables constitutive reactivity (Figure 4C) and responsiveness (Figure 4D) to growth promoting agents. In this model, the hypermethylation in the c2orf40 locus prevents an ultimate return of ECRG4 to the cell surface, thereby interfering with normal immunosurveillance and proinflammatory signaling.
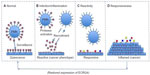  | Figure 4 A model for the action of ECRG4 in cancer and the response to injury. |
Although this model is consistent with current research, it is likely incomplete. Nevertheless, it has significant implications in the design of experiments to better understand the physiological and pathophysiological importance of ECRG4 in human biology. First, flow cytometry of ECRG4 cell surface expression might be used as a surrogate biomarker to monitor both the epigenetic state of ECRG4 gene expression and cell homeostasis. Similarly, new drugs that either regulate ECRG4 gene expression, its processing at the cell surface, or prevent hypermethylation of the ECRG4 promoter could increase the levels of cell surface ECRG4 and as such, promote the immunosurveillance and homeostasis-maintaining effects ECRG4-mediated activities.
Future directions
It is critical that the specific mechanism of ECRG4 action be identified and that the identity of the processed peptides that mediate its activities be characterized. In this way, it will be possible to address the mechanisms that regulate ECRG4 activity. Functional studies using ECRG4 gene knockout mice will enable the elucidation of ECRG4 functions when the gene is downregulated, as in after injury and in cancer. To date, knockout mice appear to have an impaired wound healing phenotype (Eliceiri BP, personal communication), but are otherwise normal. This is expected in view of the proposed sentinel function for ECRG4 expression.35,37–40 It is interesting to speculate that the loss of ECRG4 will increase susceptibility to spontaneous cancers and increase aberrant inflammatory responses to injury. If such links are identified, the findings would support a causative link between the naturally occurring downregulation of ECRG4 expression that is observed after hypermethylation and the development, progression, and metastasis of epithelial and hematopoietic cancers. These data would also suggest the possibility that ECRG4 overexpression could provide protection against cell transformation. With a better understanding of its receptor(s) and downstream signaling pathway(s), the emerging functions of ECRG4 could point to significant new therapies for human malignancies that target its physiological function at the interface of inflammation, infection, and malignancy.
Disclosure
The authors report no conflicts of interest in this work.
References
Clark HF, Gurney AL, Abaya E, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13(10):2265–2270. | |
Grimmond SM, Miranda KC, Yuan Z, et al; RIKEN GER Group; GSL Members. The mouse secretome: functional classification of the proteins secreted into the extracellular environment. Genome Res. 2003;13(6B):1350–1359. | |
Mirabeau O, Perlas E, Severini C, et al. Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res. 2007;17(3):320–327. | |
Su T, Liu H, Lu S. [Cloning and identification of cDNA fragments related to human esophageal cancer]. Zhonghua Zhong Liu Za Zhi. 1998;20(4):254–257. Chinese. | |
Bi MX, Han WD, Lu SX. Using Lab On-line to Clone and Identify the Esophageal Cancer Related Gene 4. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai). 2001;33(3):257–261. | |
Delfino KR, Southey BR, Sweedler JV, Rodriguez-Zas SL. Genome-wide census and expression profiling of chicken neuropeptide and prohormone convertase genes. Neuropeptides. 2010;44(1):31–44. | |
Svensson M, Sköld K, Nilsson A, Fälth M, Svenningsson P, Andrén PE. Neuropeptidomics: expanding proteomics downwards. Biochem Soc Trans. 2007;35(Pt 3):588–593. | |
Tegge AN, Southey BR, Sweedler JV, Rodriguez-Zas SL. Comparative analysis of neuropeptide cleavage sites in human, mouse, rat, and cattle. Mamm Genome. 2008;19(2):106–120. | |
Xie F, London SE, Southey BR, et al. The zebra finch neuropeptidome: prediction, detection and expression. BMC Biol. 2010;8:28. | |
Su H, Hu N, Shih J, et al. Gene expression analysis of esophageal squamous cell carcinoma reveals consistent molecular profiles related to a family history of upper gastrointestinal cancer. Cancer Res. 2003; 63(14):3872–3876. | |
Yue CM, Deng DJ, Bi MX, Guo LP, Lu SH. Expression of ECRG4, a novel esophageal cancer-related gene, downregulated by CpG island hypermethylation in human esophageal squamous cell carcinoma. World J Gastroenterol. 2003;9(6):1174–1178. | |
Mori Y, Ishiguro H, Kuwabara Y, et al. Expression of ECRG4 is an independent prognostic factor for poor survival in patients with esophageal squamous cell carcinoma. Oncol Rep. 2007;18(4):981–985. | |
Götze S, Feldhaus V, Traska T, et al. ECRG4 is a candidate tumor suppressor gene frequently hypermethylated in colorectal carcinoma and glioma. BMC Cancer. 2009;9:447. | |
Li LW, Yu XY, Yang Y, Zhang CP, Guo LP, Lu SH. Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int J Cancer. 2009;125(7):1505–1513. | |
Vanaja DK, Ehrich M, Van den Boom D, et al. Hypermethylation of genes for diagnosis and risk stratification of prostate cancer. Cancer Invest. 2009;27(5):549–560. | |
Li LW, Yang Y, Li XY, Guo LP, Zhou Y, Lu SX. [Tumor-suppressing function of human esophageal cancer related gene 4 in esophageal squamous cell carcinoma]. Zhonghua Yi Xue Za Zhi. 2010;90(38):2713–2717. Chinese. | |
Li W, Liu X, Zhang B, et al. Overexpression of candidate tumor suppressor ECRG4 inhibits glioma proliferation and invasion. J Exp Clin Cancer Res. 2010;29:89. | |
Li LW, Yu XY, Li XY, Guo LP, Zhou Y, Lu SX. [Mechanism of loss of human esophageal cancer-related gene 4 (ECRG4) gene expression in esophageal squamous cell carcinoma cell line EC9706]. Zhonghua Zhong Liu Za Zhi. 2011;33(8):570–573. Chinese. | |
Sabatier R, Finetti P, Adelaide J, et al. Down-regulation of ECRG4, a candidate tumor suppressor gene, in human breast cancer. PLoS One. 2011;6(11):e27656. | |
Jiang CP, Wu BH, Wang BQ, et al. Overexpression of ECRG4 enhances chemosensitivity to 5-fluorouracil in the human gastric cancer SGC-7901 cell line. Tumour Biol. 2013;34(4):2269–2273. | |
Lu J, Wen M, Huang Y, et al. C2ORF40 suppresses breast cancer cell proliferation and invasion through modulating expression of M phase cell cycle genes. Epigenetics. 2013;8(6):571–583. | |
Xu T, Xiao D, Zhang X. ECRG4 inhibits growth and invasiveness of squamous cell carcinoma of the head and neck in vitro and in vivo. Oncol Lett. 2013;5(6):1921–1926. | |
Fujimaki T, Tachibana T, Nakamura S, Murakami A, Funaki T, Kanai A. Screening of candidate genes for corneal endothelial dystrophy in the rabbit corneal endothelial cDNAs. Invest Ophthalmol Vis Sci. 2002;43:Abs 1719. | |
Nobuhiko T, Fujimaki T, Shinji N, Toshinari F, Akira M. Gene expression profile study of corneal endothelium. Juntendo Medical Journal. 2002;48(2):216–225. | |
Steck E, Breit S, Breusch SJ, Axt M, Richter W. Enhanced expression of the human chitinase 3-like 2 gene (YKL-39) but not chitinase 3-like 1 gene (YKL-40) in osteoarthritic cartilage. Biochem Biophys Res Commun. 2002;299(1):109–115. | |
Padhi BK, Joly L, Tellis P, et al. Screen for genes differentially expressed during regeneration of the zebrafish caudal fin. Dev Dyn. 2004;231(3):527–541. | |
Nishikawa M, Drmanac RT, Lobal I, Tang YT, Lee J, Stache-Crain B, Inventor; Nuvelo, Inc., San Carlos California, assignee. Polypeptide having an activity to support proliferation or survival of hematopoietic stem cell and hematopoietic progenitor cell, and DNA coding for the same. US patent 73208802008. 2008 October 21. | |
Huh YH, Ryu JH, Shin S, et al. Esophageal cancer related gene 4 (ECRG4) is a marker of articular chondrocyte differentiation and cartilage destruction. Gene. 2009;448(1):7–15. | |
Gu B, Zhang J, Wang W, et al. Global expression of cell surface proteins in embryonic stem cells. PLoS One. 2010;5(12):e15795. | |
Kujuro Y, Suzuki N, Kondo T. Esophageal cancer-related gene 4 is a secreted inducer of cell senescence expressed by aged CNS precursor cells. Proc Natl Acad Sci U S A. 2010;107(18):8259–8264. | |
Schutzer SE, Liu T, Natelson BH, et al. Establishing the proteome of normal human cerebrospinal fluid. PLoS One. 2010;5(6):e10980. | |
Tadross JA, Patterson M, Suzuki K, et al. Augurin stimulates the hypothalamo-pituitary-adrenal axis via the release of corticotrophin-releasing factor in rats. Br J Pharmacol. 2010;159(8):1663–1671. | |
Yi H, Xue L, Guo MX, et al. Gene expression atlas for human embryogenesis. FASEB J. 2010;24(9):3341–3350. | |
Farrah T, Deutsch EW, Omenn GS, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10(9):M110.006353. | |
Podvin S, Gonzalez AM, Miller MC, et al. Esophageal cancer related gene-4 is a choroid plexus-derived injury response gene: evidence for a biphasic response in early and late brain injury. PLoS One. 2011;6(9):e24609. | |
Song MH, Choi SY, Wu L, et al. Pou3f4 deficiency causes defects in otic fibrocytes and stria vascularis by different mechanisms. Biochem Biophys Res Commun. 2011;404(1):528–533. | |
Baird A, Coimbra R, Dang X, et al. Cell surface localization and release of the candidate tumor suppressor Ecrg4 from polymorphonuclear cells and monocytes activate macrophages. J Leukoc Biol. 2012;91(5):773–781. | |
Shaterian A, Kao S, Chen L, et al. The candidate tumor suppressor gene Ecrg4 as a wound terminating factor in cutaneous injury. Arch Dermatol Res. 2013;305(2):141–149. | |
Kurabi A, Pak K, Dang X, et al. Ecrg4 attenuates the inflammatory proliferative response of mucosal epithelial cells to infection. PLoS One. 2013;8(4):e61394. | |
Gonzalez AM, Podvin S, Lin SY, et al. Ecrg4 expression and its product augurin in the choroid plexus: impact on fetal brain development, cerebrospinal fluid homeostasis and neuroprogenitor cell response to CNS injury. Fluids Barriers CNS. 2011;8(1):6. | |
Tomarev SI, Wistow G, Raymond V, Dubois S, Malyukova I. Gene expression profile of the human trabecular meshwork: NEIBank sequence tag analysis. Invest Ophthalmol Vis Sci. 2003;44(6):2588–2596. | |
Vallender TW, Lahn BT. Improved DNA methylation analysis via enrichment of demethylated cells expressing an X-inactivated transgene. Biotechniques. 2006;41(4):461–466. | |
Dowell JA, Johnson JA, Li L. Identification of astrocyte secreted proteins with a combination of shotgun proteomics and bioinformatics. J Proteome Res. 2009;8(8):4135–4143. | |
Ozawa A, Lick AN, Lindberg I. Processing of proaugurin is required to suppress proliferation of tumor cell lines. Mol Endocrinol. 2011;25(5):776–784. | |
Dang X, Podvin S, Coimbra R, Eliceiri B, Baird A. Cell-specific processing and release of the hormone-like precursor and candidate tumor suppressor gene product, Ecrg4. Cell Tissue Res. 2012;348(3):505–514. | |
Podvin S, Roberton A, Johanson C, Stopa E, Eliceiri BP, Baird A. Augrin, Ecilin and Argilin: Characterization of neuropeptide candidates encoded by the esophageal cancer related Gene-4 (ecrg4) and their localization in the mouse choroid plexus. Society for Neuroscience 2009 Online. October 17–21, 2009;2:85.88/CC32. | |
Roberton A, Gonzalez AM, Stopa E, et al. Immunohistochemical evidence that argilin, the product of the ECRG4 gene, encodes a novel neuroendocrine peptide. Endocrine Abstracts. 2009;19:OC16. | |
Gonzalez AM, Roberton A, Podvin S, et al. Immunohistochemical evidence that argilin, a product of the ECRG4 gene, is a novel neurohypophyseal neuroendocrine peptide. Paper presented at: 92nd Meeting of The Endocrine Society; Jun 2010; San Diego, CA, USA. | |
Shaterian A, Kao S, Cauvi DM, et al. Identification of Ecrg4 and its cytokine-like protein product in lung: differential down regulation after exploratory laparotomy. J Surg Res. 2013;179(2):339. | |
Southey BR, Rodriguez-Zas SL, Sweedler JV. Characterization of the prohormone complement in cattle using genomic libraries and cleavage prediction approaches. BMC Genomics. 2009;10:228. | |
Omenn GS, States DJ, Adamski M, et al. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core data set of 3020 proteins and a publicly-available database. Proteomics. 2005;5(13):3226–3245. | |
Li LW, Li YY, Li XY, Zhang CP, Zhou Y, Lu SH. A novel tumor suppressor gene ECRG4 interacts directly with TMPRSS11A (ECRG1) to inhibit cancer cell growth in esophageal carcinoma. BMC Cancer. 2011;11:52. | |
Matsuzaki J, Torigoe T, Hirohashi Y, et al. Expression of ECRG4 is associated with lower proliferative potential of esophageal cancer cells. Pathol Int. 2013;63(8):391–397. | |
Nakamura Y. Retraction. Cloning and expression of soluble recombinant human esophageal cancer related gene 4 protein and its inhibitory effect on tumor growth in vitro and in vivo in esophageal carcinoma. Cancer Sci. 2011;102(6):1245. | |
Stambolsky P, Tabach Y, Fontemaggi G, et al. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell. 2010;17(3):273–285. | |
Retraction: The candidate tumor suppressor gene ECRG4 inhibits cancer cells migration and invasion in esophageal carcinoma. J Exp Clin Cancer Res. 2011;30:19. | |
Lee J, Dang X, Borboa A, Coimbra R, Baird A, Eliceiri BP. Thrombin-processed Ecrg4 recruits myeloid cells and induces anti-tumorigenic inflammation. Neuro Oncol. In press 2014. | |
Roan F, Bell BD, Stoklasek TA, Kitajima M, Han H, Ziegler SF. The multiple facets of thymic stromal lymphopoietin (TSLP) during allergic inflammation and beyond. J Leukoc Biol. 2012;91(6):877–886. | |
Ziegler SF, Roan F, Bell BD, Stoklasek TA, Kitajima M, Han H. The biology of thymic stromal lymphopoietin (TSLP). Adv Pharmacol. 2013;66:129–155. | |
Verschuere T, Toelen J, Maes W, et al. Glioma-derived galectin-1 regulates innate and adaptive antitumor immunity. Int J Cancer. 2014;134(4):873–884. | |
Woo JM, Park SJ, Kang HI, et al. Characterization of changes in global gene expression in the brain of neuron-specific enolase/human Tau23 transgenic mice in response to overexpression of Tau protein. Int J Mol Med. 2010;25(5):667–675. | |
Camões MJ, Paulo P, Ribeiro FR, et al. Potential downstream target genes of aberrant ETS transcription factors are differentially affected in Ewing’s sarcoma and prostate carcinoma. PLoS One. 2012;7(11):e49819. | |
Matsuzaki J, Torigoe T, Hirohashi Y, et al. ECRG4 is a negative regulator of caspase-8-mediated apoptosis in human T-leukemia cells. Carcinogenesis. 2012;33(5):996–1003. | |
Li L, Zhang C, Li X, Lu S, Zhou Y. The candidate tumor suppressor gene ECRG4 inhibits cancer cells migration and invasion in esophageal carcinoma. J Exp Clin Cancer Res. 2010;29:133. | |
velocigene.com [database on the Internet]. International Knockout Mouse Consortium. Available from: http://www.velocigene.com/komp/detail/10814. Accessed October 14, 2014. | |
Gene Tools [database on the Internet]. Available from: http://www.genetools.us/. Accessed March, 2013. | |
Promoter Prediction Server [database on the Internet]. Lyngby, Denmark: Technical University of Denmark. Available from: http://www.cbs.dtu.dk/. Accessed March, 2013. | |
FPROM [database on the Internet]. Mount Kisco, NY: Softberry, Inc. Available from: http://linux1.softberry.com/. Accessed March, 2013. | |
e!ENSEMBL [database on the Internet]. Hinxton, UK: ENSEMBL. Available from: http://www.ensembl.org/. Accessed August, 2014. | |
Clustal Omega [database on the Internet]. Dublin: Conway Institute UCD Dublin. Available from: http://www.clustal.org/omega/. Accessed March, 2013. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
