Back to Journals » Clinical and Experimental Gastroenterology » Volume 12
Esophageal cancer genetics in South Africa
Authors Alaouna M , Hull R , Penny C, Dlamini Z
Received 7 August 2018
Accepted for publication 7 January 2019
Published 23 April 2019 Volume 2019:12 Pages 157—177
DOI https://doi.org/10.2147/CEG.S182000
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Andreas M. Kaiser
Mohammed Alaouna,1 Rodney Hull,2 Clement Penny,1 Zodwa Dlamini2
1Department of Internal Medicine, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa; 2Research, Innovation & Engagements Portfolio, Mangosuthu University of Technology, Durban, South Africa
Abstract: Esophageal cancer (EC) is an extremely aggressive cancer with one of the highest mortality rates. The cancer is generally only diagnosed at the later stages and has a poor 5-year survival rate due to the limited treatment options. China and South Africa are two countries with a very high prevalence rate of EC. EC rates in South Africa have been on the increase, and esophageal squamous cell carcinoma is the predominant subtype and a primary cause of cancer-related deaths in the black and male mixed ancestry populations in South Africa. The incidence of EC is highest in the Eastern Cape Province, especially in the rural areas such as the Transkei, where the consumption of foods contaminated with Fusarium verticillioides is thought to play a major contributing role to the incidence of EC. China is responsible for almost half of all new cases of EC globally. In China, the prevalence of EC varies greatly. However, the two main areas of high prevalence are the southern Taihang Mountain area (Linxian, Henan Province) and the north Jiangsu area. In both countries, environmental toxins play a major role in increasing the chance that an individual will develop EC. These associative factors include tobacco use, alcohol consumption, nutritional deficiencies and exposure to environmental toxins. However, genetic polymorphisms also play a role in predisposing individuals to EC. These include single-nucleotide polymorphisms that can be found in both protein-coding genes and in non-coding sequences such as miRNAs. The aim of this review is to summarize the contribution of genetic polymorphisms to EC in South Africa and to compare and contrast this to the genetic polymorphisms observed in EC in the most comprehensively studied population group, the Chinese.
Keywords: esophageal squamous cell carcinoma, ESCC, adenocarcinoma, alcohol, smoking, diet, South Africa, China
Introduction
Esophageal cancer (EC) is a malignant tumor in the epithelial cells padding the esophagus.1 Worldwide, EC is the third most frequently diagnosed malignancy of the upper digestive tract. EC is responsible for >400,000 deaths each year, making it the seventh leading cause of cancer-related deaths.2–4 The regions with the highest number of cases and most deaths are the south and the east of Africa, central Asia, Turkey, Iran, Kazakhstan and China (Figure 1).5 The disease has a poor prognosis with a 5-year survival rate of <10%.6 The two most important histological forms of EC are etiologically and pathologically unrelated. These are esophageal squamous cell carcinoma (ESCC), which predominates in non-white populations, and adenocarcinoma, which is more common in white population groups.4,7,8 EC occurrence and type are strongly influenced by the geographic area and ethnicity.4,7,8 Most incidences of esophageal malignancy are ESCC, which makes up >95% of all cases.9 However, the number of adenocarcinoma cases is on the rise, with the proportion of adenocarcinomas increasing from 3.5% in 1985 to 17.0% in the year 2000. This is especially true in western countries, where it now accounts for 30%–50% of all diagnosed ECs.10–12 One of the main environmental factors that has been investigated as an associative factor contributing to the development of ESSC is nutrition. Nutritional factors that may play a role in the development of ESSC include a diet low in dietary vitamins and minerals, the consumption of hot beverages and pickled food and exposure to foods contaminated with or containing nitrosamines.13,14 However, while these environmental risk factors may be factors that strongly contribute to the development of ESCC, they alone may require a genetic component which would predispose an individual to ESCC.4,15
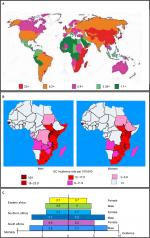  | Figure 1 Incidence and mortality rates of EC. Notes: (A) Worldwide in both sexes and (B) in men and women in Africa. The highest incidence rate is in South Africa and East Africa, with the highest incidence in both sexes being found in Kenya, Uganda and Malawi. A high incidence rate of EC is found in both sexes in Guinea Bissau. (C) The incidence and mortality rates per 100,000 in South Africa and the Eastern Africa and Southern Africa regions is much higher in males than in females.4 Abbreviation: EC, esophageal cancer. |
Cancer of the esophagus was an uncommon disease in South African during the 1920s and 1930s.16 This was followed by a rapid rise in the number of diagnosed cases of the disease.17–19 EC has become the third most commonly diagnosed cancer in Black South Africa. Age occurrence values are 22.3 per 100,000 in black males and 11.7 per 100,000 in black females, respectively.20 It is also the fourth highest cause of death in males of the colored or mixed ancestry group.21 EC rates increase noticeably in South Africa between the average age of 60 and 70 years, along with cigarette fuming and extreme alcohol use (Table 1).22 The Transkei region is considered as the center of the malady in South Africa,23 with a standard incidence rate of 46.7/100,000 for males and 19.2/100,000 for females.24 The increase in EC rates in South Africa over the last decades can largely be attributed to changes in environmental factors and exposure to carcinogens.23 Two of the most important factors are increased alcohol intake (Table 1)16 and increased tobacco use (Table 1).25 However, other weak carcinogens also play a role, such as the exposure to environmental smoke26 mainly in the form of cooking fires, the use of wild herbs such as Solanum nigrum, human papillomavirus (HPV) infection23 and the consumption of traditional beer brewed with maize that may contain fungal mycotoxins or nitrosamines.16 Another important factor contributing to the increase in EC rates is the adoption of a western diet. This includes increased consumption of fats and animal protein as well as a decrease in the vitamin content of traditional beer that is now brewed almost exclusively with maize.16
  | Table 1 Alcohol consumption and tobacco use in China and South Africa Notes: The amount of pure alcohol consumed per capita and the prevalence of tobacco use in China and South Africa are presented in the above table. For alcohol consumption, data are shown for the periods 2003–2005 and 2008–2010 and for males and females in 2010. The trend observed in both countries is an increase in alcohol consumption over time.187 In terms of tobacco use, data are given as percentages of the population that uses tobacco. Abbreviations: ASIRW, age-standardized incidence rate by world standard population. |
Another region of the world with a high incidence rate of EC is China. Over half of all new cases of EC are diagnosed in China. In South Africa, EC is the eighth most common cancer in men and the eleventh in women, while in China, it is the fourth most frequently diagnosed cancer.4 Like South Africa, the areas in China with the highest incidence rates are the rural areas.27 Areas such as Henan, Hebei and Shanxi in Central North China have the highest incidence rates in the world (over 100 per 100,000).28,29 Additionally, the provinces of Sichuan, Anhui, Jiangsu, Hubei, Fujian, Guangdong and Xinjiang have age-adjusted rates >30 per 100,000 (Figure 2).30 Overall, EC is the fourth leading cause of cancer-related deaths in China.4 However, in Xi’a city in Shaanxi Province, it is the leading cause of cancer-associated mortality with a mortality rate of 24 per 100,000.31 Once again, environmental factors similar to those suspected of playing a role in South African cases of EC are also thought to be important contributing factors in China. These include nutritional factors such as the consumption of fatty meat, salted and pickled vegetables and moldy food, as well as nutritional deficiencies. Lifestyle factors include passive smoking, esophageal lesions, infection with Helicobacter pylori, low socioeconomic status and poor oral hygiene. However, a family history of cancer is also an important risk factor associated with an increased risk of ESCC,32 and this points to a genetic component.
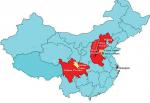  | Figure 2 Geographic locations of the four high-risk areas in China. Notes: The counties Cixian, Shexian and Linxian are located on the borders of the Henan and Hebei provinces. These three counties are located in an area with a radius of 60 km. The Taihang Mountains straddle the border of the Shanxi and Hebei provinces and have some of the highest mortality rates of esophageal cancer in the world. Another high-risk area for esophageal cancer is Yanting in Sichuan Province. Data from Lin et al.37 |
Despite the known dominant role played by environmental factors in increasing the rates of EC, the occurrence of high rates of EC in members of the same blood family observed in both China and South Africa suggests that genetics may also play an important role.26
Environmental risk factors for EC
Environmental risk factors for EC are complex and have a complicated relationship with genetic and etiological elements concerning the development of ESCC and adenocarcinoma.33 The influence of these different factors on the risk of developing adenocarcinoma and ESCC is given in Table 2. The geographic variation in the occurrence of EC is likely caused by genetic polymorphism and unstable environmental factors.6 Worldwide, the primary risk factors for ESCC are alcohol use and tobacco smoking. Other factors rely on the region of the world. These include hot brews in Iran and South America, and smokeless tobacco and alimentary deficits in China, Central Asia and South Africa.34 Other risk factors include the extensive utilization of wood and charcoal for cooking and heating,35,36 along with nutritional factors that are related to the growth of the EC.6
  | Table 2 Comparison of the risk factors for adenocarcinoma and ESCC Abbreviations: BMI, body mass index; ESCC, esophageal squamous cell carcinoma. |
Analyses carried out within the areas of the Transkei region and KwaZulu-Natal Province have demonstrated that people at highest risk are commonly poorer, have inadequate diets and consume some class of alcohol and tobacco.31,32,37 Furthermore, bacteria from the family Helicobacteraceae were discovered in roughly 50% of South African patients with EC.38 The use of any form of tobacco was found to increase the risk of developing EC by as much as four times in the Eastern Cape Province of South Africa.39 Currently, the prevalence of tobacco consumption among adult South Africans is 17.6%, with males being four times more likely (29.2%) than females (7.3%) to use tobacco products.40 Cigarette smoke is composed of over 4,000 chemical compounds, and >60 of these are recognized to cause diseases, including cadmium, nicotine/cotinine and benzo[a]pyrene.41 In South Africa, the consumption of >53 g of ethanol per day led to a risk of developing EC five times greater in comparison to non-drinkers, while a combination of both alcohol, consumption and smoking led to an 8.5 times greater risk of developing EC. The attributable fractions in high-risk populations in South Africa were 58% for smoking, 48% for alcohol consumption and 64% for both factors combined.39 In comparison, tobacco use in China is much higher in Chinese males than females. Currently, the prevalence of smoking in China is 56.1% among males and 2.2% among females in rural areas. In urban regions, the occurrence was 49.2% among males and 2.6% among females.42 The attributable fraction for both tobacco and alcohol consumption was lower in high-risk Chinese populations compered to their South African counterparts. The attributable fractions of esophageal patients who were smokers were 17.9% in Chinese men and 1.9% in Chinese women,43 and for alcohol consumption they were 15.2% in Chinese men and 1.3% in Chinese women.44 This higher tobacco use and alcohol consumption among males in both South Africa and China (Table 1) is most likely the major factor contributing to the higher incidence of EC observed in males.
In the Eastern Cape of South Africa, an increase in the risk of developing EC is associated with a diet low in green leafy vegetables and fruit.45 When examining the diet of those in high-risk, remote rural areas of South Africa, it was found that low plasma concentrations of vitamins A, E, B12, folic acid and selenium were also associated with an increased risk of developing EC.46 Similarly, reports in Linxian, China showed that general undernourishment, as well as deficiencies in selenium, zinc, folate, riboflavin and vitamins A, C, E and B12 were linked with amplified risk of ESCC.47 Another dietary cause of EC in South Africa is the production of carcinogenic nitrosamines resulting from fungal contamination of food. The shift in the diet of the black population of South Africa from sorghum to maize is one of the factors that have been identified as contributing to the current high levels of EC in this population group. Unlike sorghum, maize is frequently contaminated with fungi such as Fusarium verticillioides. These fungi produce toxins such as fumonisins, which reduce nitrates to nitrites and synthesize nitrosamines.48 Homemade traditional beer is commonly consumed in these regions of South Africa. This beer was once brewed mainly from sorghum, but is now generally brewed from maize. This traditional beer has been found to contain high amounts of carcinogenic N-nitrosamines.49 The maize-rich diet also leads to high levels of prostaglandin E2 (PGE2) in gastric fluid. This enzyme activates the Wnt signaling pathway. PGE2 also stimulates proliferation and may predispose esophageal cells exposed to gastric fluid to developing ESCC.50 Linoleic acid is used to synthesize prostaglandins, and therefore, high levels of linoleic acid lead to increased PGE2 activity in the stomach.51 PGE2 represses gastric acid secretion and leads to a reduction in the tone of the muscles that control pyloric and lower esophageal sphincters. This results in chronic hyperchlorhydria duodenogastroesophageal reflux (DGOR).52 Chronic alkali DGOR is thought to lead to the denaturation of proteases due to the altered pH. The inhibition of proteases leads to increased growth factor activity as they are no longer broken down by proteases. This leads to an increase in the risk of developing ESCC.53 The cyclooxygenase enzyme (COX-2) is required to convert a linoleic acid derivative into PGE. Like PGE2, the level of COX-2 is elevated in EC.36,54–57 It has previously been confirmed that gastric fluid out of specimens from Transkei contained higher levels of PGE2.50 In China, analysis was performed on local sources of drinking water and samples and in food samples, in order to determine the level of nitrates and nitrites in these samples. It was determined that these concentrations were abnormally high and, therefore, indicated that high levels of nitrosamine were present.58 Another common causative agent is infection with H. pylori and HPV. However, case studies in South Africa found no association between EC and an increase in H. pylori infection.59 However, in Chinese populations, the relationship between H. pylori infection and ESCC was significant.60 This shows an important difference between environmental factors that may lead to ESCC in Chinese and western populations. In terms of HPV infection and its relationship to EC in South Africa, HPV DNA could only be isolated from 9% of ESCC patients. HPV infection appears to only play a minor role in EC.61 However, looking for HPV biomarkers (E6, E8 and p16) in ESCC tissue samples from China led to the conclusion that there was no association between HPV infection and ESCC.62
Epidemiologic studies in Chinese populations have studied the link between EC and other factors such as the consumption of warm beverages in the form of green tea. While there seemed to be no relationship between drinking tea and ESCC, either directly or inversely, there was a relationship between drinking hot green tea and the development of ESCC. This implies that consumption of any high-temperature beverage could increase the chances of developing ESCC.63 Another factor that has been studied is gastric atrophy. This refers to dying of gastric cells and their replacement with intestinal tissues. Gastric atrophy is indicated by lower serum ratios of pepsinogen I/pepsinogen II. In Chinese populations, gastric atrophy increases the risk of ESCC as indicated by lower Pepsinogen I to Pepsinogen II ratios being positively associated with ESCC.13
These risk factors have the capacity to influence the stability of fragile sites.64 Tobacco amplifies the expression of regular sensitive locations;65 HPV fuses within fragile site loci.66 Alcohol and fumonisin each influence folate levels, which facilitates the expression of fragile sites.64,67,68 The addition of fumonisin to cell cultures increases the frequency of chromosomal damage.69 The combination of these risk factors leads to the initiation of genetic instability at the early stages of carcinogenesis, with the fragile sites being prime targets.64 It would be of great benefit, for diagnostic and prognostic purposes, if the genes within the most common fragile sites could be determined in South African cases of ESCC.64
Genetic polymorphisms
The importance of genetic factors in contributing to increased risk of developing ESCC was demonstrated by a study undertaken in the Tenwek Mission Hospital in Western Kenya. Kenya is a very high-risk area for ESCC and is part of the ESCC corridor (Figure 1). In this study, ESCC patients younger than 30 were found to have a high incidence of cancer within their family with 20 out of 60 patients having a family history of EC. All these patients also originated from the same community (Kipsigis).70 Many of the genes that have been linked to the development of EC71,72 were found to be associated with DNA maintenance and repair, alcohol folate and carcinogen metabolism, cell cycle regulation and apoptosis.73 Nevertheless, just a few of these genes have shown to be associated with disease vulnerability.72 Therefore, some additional genetic factors assist in predisposing individuals to EC.71 The most common genetic polymorphisms are single-nucleotide polymorphisms (SNPs). A large percentage of the genetic variations that occur in the human genome are due to SNPs.74
A xenobiotic is any chemical substance that is foreign to an organism in its normal physiological function. Humans are exposed to multiple environmental conditions that may result in the buildup of xenobiotics within the body tissues. Many dietary and environmental xenobiotics would require metabolic activation by the Phase I or Phase II enzymes to exert their carcinogenic effect.75 The Phase I enzymes catalyze the conversion of a toxic or insoluble compound into a polar, water-soluble metabolite through oxidation. The resulting metabolites contain functional groups such as OH, NH2 and SH. These groups are sites for reactions catalyzed by the Phase II enzymes.75 The Phase II enzymes give rise to compounds that are more hydrophilic and thus easily excreted in the urine.75 Although the Phase I and Phase II enzymes act in the detoxification pathway, they usually generate unstable and more reactive intermediate compounds that, if not quickly removed, can bind covalently to DNA and generate DNA adducts with mutagenic and/or carcinogenic properties.76 The presence of xenobiotics and polymorphisms in xenobiotic-metabolizing enzymes (XMEs) can increase the risks of sensitivity to ESCC.77 These XMEs include Phase I enzymes such as CYP1A1, CYP1B1, CYP2A6 and CYP2E1 and Phase II enzymes such as GSTM1, GSTT1 and GSTP1 (Table 3).31 Variations at 10q23 in PLCE1 were linked with ESCC (Table 3).78 Alteration in the riboflavin transporter C20 ORF54 on chromosome 20p13 is a risk factor for ESCC.43 ESCC is related to polymorphisms of the ADH enzymes ALDH2 and ADH1B1. These enzymes are engaged in alcohol metabolism, and with CYP1A1, are involved in xenobiotics’ detoxification.79 NQO1 is one of the enzymes belonging to the NAD(P)H dehydrogenase (quinone) family that are involved in protecting cells from oxidative damage.80 A significant polymorphism dealing with a single C to T replacement at nucleotide 609 of exon 6 in the NQO1 cDNA affects the NQO1 enzyme activity that induces a Pro187Ser amino acid substitution.81 The NQO1 C609T polymorphism has been related to ECs.82–84 NQO1 is involved in cellular antioxidant defense systems by detoxifying quinines.85 A meta-analysis showed that the NQO1 C609T polymorphism considerably increases the risk of developing EC.85
The role of polymorphisms in alcohol metabolising enzymes: ADH and ALDH in the risk of developing EC
The WHO classifies alcohol as a Group 1 carcinogen. In the body, alcohol is initially metabolized by the ADHs to acetaldehyde. This is then further metabolized to acetic acid by ALDH2. Mutations and polymorphisms that affect the functioning of these enzymes may affect the ability of the individual to detoxify ethanol, leading to increased exposure of cells to carcinogens such as acetyl aldehyde which is dissolved in the saliva following consumption of alcohol or smoking tobacco (Table 4).86 There is a guanine to adenine SNP within ALDH2 at position 1510, which leads to a lysine at codon position 487. This leads to a catalytically inactive subunit, and the allele containing this polymorphism is termed ALDH2*two (wild-type ALDH*1). Individuals homozygous for this mutation (ALDH2 *1/*2) have only 6.25% of normal ALDH2 *1/*one activity.87 The ADH 1B *two mutant allele results from an SNP in Exon 3 of ADH1B, resulting in a His to Arg substitution at codon 48. This results in a protein with a much higher activity than the wild type encoded by the ADH1B *one allele. The homozygous ADH1B*2/*two polymorphism has a Vmax 40 times higher than the wild-type ADH1B*1/*1. This results in greater amounts of alcohol being oxidized by the more active enzyme.88 Both these polymorphisms are frequently found in individuals originating from East Asia. There is a strong association between the ALDH2 *2/*two genotype and EC (OR =4.42). The lower activity of ADH1B *one allele is also associated with an increase in the risk of developing EC. This occurs in a manner associated with the number of ADH 1B *one alleles present, with *1/*one homozygous having a greater risk for developing EC than the *1/*two allele. In South Africa, the role played by mutations in the genes coding for these enzymes has not been studied intensively. The ALDH2*two allele is present in black and mixed ancestry South African populations, where it is significantly associated with an increased risk for EC (OR =2.35; Table 4).89,90 The ADH1B *two allele was also detected in the mixed ancestry population in South Africa. As in other population groups, this allele is associated with a decreased ESCC risk.90 Mutations affecting the function of ALDH2 are so prevalent among East Asian population groups that it is common for individuals in these population groups to have a flushed skin or red blotches on their face, neck and shoulders following consumption of alcohol. This is known as the alcohol flush reaction. People displaying this reaction have an increased risk of developing EC. This rash results from excess acetyl aldehyde, with the polymorphism most commonly associated with the flush response being the rs671 allele of ALDH2. This mutation in ALDH2 and the flush reaction occur in 30%–50% of South East Asians, but it is extremely rare in Europeans and sub-Saharan Africans (Tanzania =0.0025%). In South Africa, it is, therefore, of no use as a diagnostic or predictive tool in assessing the susceptibility of an individual to alcohol-related risks of developing ESCC.91
The role of genetic polymorphisms in the androgen receptor (AR) gene in the risk of developing EC
The AR is a nuclear receptor that translocates to the nucleus and acts as a transcription factor once it has bound its ligand. This ligand is generally any of the androgenic hormones. As a transcription factor, it controls pathways associated with cellular proliferation and differentiation.92 The receptor is expressed in multiple cancer tissues and cell lines. These include neoplastic colon tissues,93 breast tumors,94 hepatocellular cancer95 and ESCC.96 AR is a part of the steroid receptor family and the AR gene is situated at q11-12 on the X chromosome.97 The AR gene is 75,000–90,000 nucleotides in length, consisting of eight exons.98 The various protein isoforms range in size from 910 to 919 amino acids and consist of four domains. The first is an NH2-terminal transactivation domain. A central C4 zinc finger DNA-binding domain and a nuclear-localizing short hinge region follow this. Finally, the C terminal consists of a steroid hormone ligand-binding domain.99 The gene for the AR is highly polymorphic in human populations due to the fact that the first exon contains two polymorphic poly amino acid tracts, a glutamine (CAG) 7–31 repeat96,100,101 and a glycine (GGC)102–105 8–20 repeat106,107 ~1.1 kb apart. Examination of data concerning the (GGC) n allele indicates a genetic sensitivity element for EC in black males. A short (GGC) n allele was involved in the disease in this population, with a GGC shorter than 16 being associated with the disease with an OR of 2.7, 95% CI, and a GGC shorter than 14 being associated with EC with an OR of 3.3, 95% CI.108 Among the Chinese population in Beijing, shorter CAG repeats were observed in male EC patients, implicating this polymorphism in predisposing individuals to a greater risk of developing EC.109
The role of genetic polymorphisms in glutathione S-transferase (GST) genes in increased EC risk
The GSTs are Phase I enzymes that introduce reactive or polar groups onto a compound as the initial step to detoxify or remove a reactive or dangerous compound. GSTs accomplish this by catalyzing the conjugation of glutathione (GSH) to electrophilic compounds.110 These compounds include carcinogens that may lead to the development of cancers such as ESCC.
The risk of developing ESCC in South African and Chinese populations is a result of genetic polymorphisms in an individual’s GST-coding genes and their exposure to environmental factors such as tobacco smoke and alcohol consumption (Table 5).71 There are four major subfamilies of human GST genes, which are divided into the categories GST A, M, T and P.111 Deletion polymorphisms have been observed in two of these families, GSTM1 and GSTT1.112 Polymorphisms of GSTM 1, GSTT1 and GSTP1 have been demonstrated to be linked to susceptibility to various forms of cancer. This is especially true concerning cancer triggered by exposure to carcinogens such as cigarette smoke,113 and they also play a role in the resistance to chemotherapy treatment114 and in disease outcomes.115
GSTP1 is expressed in hepatic tissues, lungs and the esophagus and at very low levels in the liver.111 Moreover, it has been shown to be overexpressed in several malignant tissues compared to normal tissues. Polymorphisms of GSTP1 take part in EC within a pathway of malfunctions in the p53 malignant tumor suppressor gene, which is prevalent in ESCC.116 The human GSTM1 gene is composed of seven exons, is 5.9 kb in length and is found on chromosome 1p13.117 The human GSTT2 gene, located on chromosome 22q11.2, is made up of five exons spanning 3.8 kb in length.118
miRNA polymorphisms in EC development
Altered miRNA transcription changes the expression of oncogenes and tumor suppressor genes. These genes affect processes such as proliferation, apoptosis, as well as the motility and invasiveness of cancer cells. This has been found to apply to ESCC as well.119–121 Different studies have identified different numbers of miRNA whose transcription levels change in ESCC. Ogawa et al reported that 22 miRNAs were upregulated in ESCC tissue, while 4 miRNAs were downregulated.122 Yao et al found 27 downregulated and 16 upregulated miRNAs in ESCC tissue, while Ren et al detected 51 upregulated and 17 downregulated miRNAs.123 Next-generation sequencing has identified 78 diversely expressed miRNAs in ESCC.124 Silencing of cancer genes mainly occurs through DNA methylation. DNA methylation can, therefore, affect a range of cellular processes. These include the regulation of the cell cycle checkpoint, apoptosis, signal transduction, cell adhesion and angiogenesis. Hence, DNA methylation has a role in the deregulation of miRNAs in cancer, and miR-145, miR-30a-3p, miR133a and miR-133b are the possible tumor promoters.125
In South Africa, the polymorphisms in miRNA genes have been connected to many cancer types along with ESCC.126,127 In black South Africans, a polymorphism in miRNA 3184, known as rs6505162, results in associated risks for ESCC. The location of the rs6505162 SNP allows it to influence two different miRNAs and a single protein coding gene. These include miR-423, miR-3184 and NSRP1. The two miRNAs, miR-423 and miR-3184, are oppositely oriented and overlap at rs6505162. The SNP is situated downstream of miR-423 and upstream of miR-3184.128 Therefore, it is more likely that rs6505162 influences the transcription of miR-3184 rather than that of miR-423.129 The increased risk of ESCC related to this SNP is also linked to environment. It was found that the rs6505162 polymorphism was related to increased cancer risk in black African patients who lived in an environment where they were subjected to high levels of smoke inhalation.128 The risk of developing ESCC was greater in those individuals who possessed the SNP and used solid fuels for cooking than in those individuals with the SNP and who used electricity or gas for cooking. Due to population heterogeneity, this relationship does not seem to be present in the mixed ancestry group.128 Two other SNPs previously identified as playing a role in EC, rs213210 (miR219-1) and rs7372209 (miR26a-1), were found to interact in both the black and mixed ancestry populations to reduce the risk of cancer development. Individuals with the genotype AArs213210-CTrs7372209 had a reduced risk of developing ESCC.128
Polymorphisms in miRNA genes related to ESCC risk have also been identified in Chinese populations.130 The SNP rs2910164 C > G in miR-146a increases the risk of individuals developing ESCC within the Han Chinese population, and the increased risk posed by the rs2910164 GG genotype was more notable in cigarette smokers.131 The rs11614913 TC polymorphism in miR-196a2 is predicted to reduce ESCC risk among females who have never smoked or consumed alcohol. However, in males or those who smoke and drink, the miR-196a2 r s11614913 TC, CC or TC/CC genotype may play a role in increasing the risk of developing ESCC.132 Similarly, individuals who did not smoke or drink alcohol and possessed the hsa-miR-34b/c rs4938723 CC genotype had a reduced threat of ESCC.130,133 The risk of developing ESCC is increased by the SNPs rs6505162 C.A in Hsa-miR-423133 and rs531564 CG SNP in pri-miR-124-1.130,134
The role of polymorphisms in the DNA mismatch repair (MMR) genes and tobacco smoking in increasing EC risk
Environmental toxins are able to cause cancer due to their ability to damage DNA. An important safeguard against this is the DNA MMR enzyme system. The MMR system restores mistakes within DNA. These mistakes take place during normal DNA metabolism; however, they are also related to cancer caused by exposure to environmental carcinogens.73 Studies suggest that the MMR gene polymorphisms control the etiology of EC. These genes are connected to the processes of DNA preservation and repair, alcohol, folate and carcinogen metabolism, cell cycle control and apoptosis.73 Furthermore, MMR genes and their polymorphisms account for the increased risk in the development of lung or head and neck cancer, including EC.36,135–138 A list of some of the polymorphisms in MMR genes that may contribute to the risk of EC is given in Table 6. The DNA MMR process is made up of MLH1, MSH2, MSH3, MSH6 and PMS2 proteins.139 During DNA synthesis, the MMR corrects the microsatellite instabilities (MSIs) in order to sustain genomic integrity.140 MSI is characterized by increased mutations in microsatellites that arise due to germline MMR gene mutations causing Lynch syndrome. MSI can also be caused through the epigenetic inactivation of the MLH1 gene and the CpG island methylator phenotype as a consequence of MLH1 hypermethylation.141 This may eventually lead to carcinogenesis due to replication errors that remain uncorrected in essential cancer regulating genes.142
Studies searching for extensive microsatellite modification in EC have discovered low-level MSI in 16%–67% of adenocarcinomas, while 2%–60% of squamous cell carcinoma tumors were MSI-L positive. These high-incidence populations possessed the maximum MSI abundances, signifying that MMR might be included in the pathogenesis of the esophagus.143,144 Genetic conversions in microsatellite regions, a property of a faulty DNA MMR procedure, have been found in ECs.73
In black South Africans, no association between MMR polymorphisms and cancer risk was observed at the SNP level. It was suggested that different linkage disequilibrium patterns are the reason for the lack of significant associations in these individuals.73 On the other hand, three regular polymorphisms were allied with ESCC in mixed ancestry South Africans.73 Epithelial cells of the esophagus are regularly exposed to DNA destructive compounds and should have the capacity to fix the DNA damage provoked by numerous carcinogens in food, tobacco and solid fuel smoke.132 A polymorphism within the DNA repair associated gene MSH3 (Table 6) may interact with cigarette in some cases of carcinogenesis. A model was developed to explain the role played by the interaction between the MSH3 protein and tobacco smoke in EC,145 as MSH3 is implicated in playing a significant role in DNA double-strand break (DSB) repair.146–148 Cigarette smoke has been shown to damage DNA by causing DSBs.149 Therefore, any mutation or polymorphism decreasing MSH3 activity may increase the risk of developing EC.
Promoter CpG island methylation appears to be a frequent event in ESCC carcinogenesis, with methylated sequences of hMLH1, hMSH2 and MGMT being identified in a large proportion of Chinese ESCC patients.150 Promoter methylation in the ML1 promoter of male Han Chinese ESCC patients is associated with a poor prognosis.151
Genetic polymorphisms of CYP genes increase the risk of developing EC
CYPs play a considerable role in detoxifying chemicals that the esophagus is exposed to.152,153 The CYP enzymes are expressed in the esophageal mucosa, suggesting that this tissue can detoxify DNA-binding chemical carcinogens.154–156 CYP2E1 is an 11.4 kb gene containing nine exons positioned on chromosome 10q26.345.157 The CYP2E1*six heterozygous genotype is linked to ESCC in South Africa158 and the c1/c1 genotype is linked to ESCC in China (Table 7). CYP3A is mostly regulated by genetic polymorphisms in the CYP3A4 and CYP3A5 genes, which exist together in a 231 kb area on chromosome 7q21.1.159 CYP3A5 is an important enzyme in the esophagus and metabolizes carcinogenic compounds,152 playing a role in both metabolic catalysis of xenobiotics to produce reactive intermediates found in the pathogenesis of EC and in the detoxification of carcinogens that the esophagus is exposed to. Functional polymorphisms in the CYP3A5 gene are mainly represented by CYP3A5*three and CYP3A5*six alleles.160 CYP3A5*six appears to be African specific.161 In South African populations, the CYP3A5*three allele was found to be present in 14%–16% of the black subjects and 48%–59% of the mixed ancestry population (Table 7).162
As mentioned previously, South African maize is contaminated with high levels of aflatoxins and fumonisin.163 Fumonisins stimulate xenobiotic metabolizing enzymes along with the CYPs.164 CYP3A5 could be slightly decreased. CYP3A enzymes metabolize several steroids such as progesterone, estradiol, testosterone and corticosterone.160 Similar associations between environmental factors and the CYP3A5*three allele have been found in a high-risk Chinese population from Shanxi Province.158
Genetic polymorphisms in genes encoding sex hormone metabolizing enzymes increase the risk for EC
Males are two to four times more likely to develop EC than females. Also, while men are more commonly exposed to risk factors, one study seemed to indicate that these factors had an even greater harmful effect on females than they did in males (Table 8).165 Evidence from epidemiological and experimental studies suggests that sex hormones may play a significant role in the development of ESCC.15 Since hormone therapy is able to lower the risk of ESCC,78,166,167 it has been suggested that low estrogen plays a role in increased risk of ESCC.168 One line of evidence that suggests this is the increased levels of estradiol, which is an estrogen receptor agonist, in patients with ESCC, further suggesting that low levels of estrogen are associated with an increased risk of ESCC.168 This implies that mutations in genes that play a role in metabolizing sex hormone may be important in the etiology of ESCC. Genetic variation in at least six of these genes that play a role in the metabolism of sex hormones have been associated with an increase in the risk of developing EC. These include cytochrome P450s (CYP1B1, CYP3A7, CYP3A5 and CYP11A1), SULT2B1 and SHBG. These genes regulate sex hormone activity by catalyzing their breakdown or regulating their bioavailability.15
  | Table 8 The extent to which different risk factors influence the odds of developing ESCC in Swedish males and females165 Abbreviation: ESCC, esophageal squamous cell carcinoma. |
Earlier reports showed that ESCC cells grown in vitro express both estrogen and ARs and the addition of these hormones to the growth media promoted the growth of ESCC cells in vitro.96,169
Estrogen is metabolized into 4-hydroxy estrogen by the enzyme CYP/CYP450 1B1 (CYP1B1). This enzyme has a high catalytic activity for this reaction. The resulting 4-hydroxy estrogen is able to induce DNA damage.170 Mutations in Cyp1B1 that result in a GG substitution rather than the GA in normal copies decrease the efficacy of the enzyme. South African individuals homozygous for the mutant allele (GG), and therefore possessing a less-efficient CYP1B1, had a lower risk of developing ESCC. These heterozygous individuals (GA) had an increased risk of developing ESCC.158
SULT2B1 catalyzes the sulfate conjugation of many hormones, and is therefore required for the formation of these hormones. Two SNPs, SULT2B1a and SULT2B1b, in this gene are connected with the risk of ESCC in the Chinese population.160
The role of dietary selenium in contributing to EC
Selenium is an essential factor in several metabolic activities. This is due to the use of selenium by selenoproteins, proteins containing a selenocysteine amino acid residue. These proteins function as enzymes in many vital processes. These include protecting the cell membranes from lipid peroxidation, defending cells from oxidative damage and regulation of the immune system.171 Selenium has anticarcinogenic and chemoprotective effects, and selenium-containing proteins perform a significant function in the metabolism and detoxification of polycyclic aromatic hydrocarbons.172 Selenium also protects the DNA from being damaged by oxygen free radicals by scavenging these oxygen molecules. Selenium also promotes the removal of damaged cells by inducing apoptosis. If they are not removed, these damaged cells could potentially develop into cancerous cells.173
Selenium compounds stimulate apoptotic death of tumor cells,174 and selenium regulates p53 in the role it plays in DNA repair or apoptosis.175 Primarily, selenium is found as selenomethionine. Selenomethionine stimulates the repair of DNA damage through the p53 pathway. An essential component of this signaling pathway is the GSTP1 protein. GSTP1 activates p53 through a redox mechanism.176 The function and activity of a mutant GSTP1 protein that has an Ile-to-Val substitution (rs947894) are decreased. This mutant protein is the result of a polymorphism in the GSTP1 gene at codon 105.177 Mutations in the p53 gene are prevalent in all cancers including EC.178,179 A single base modification from arginine (CGC) or proline (CCC) is initiated on codon 72 (rs1042522).180 This polymorphism is connected to tumorigenesis in a variety of cancers. It is also a risk factor for HPV-associated cervical neoplasia and ESCC.181,182 The GSTP1 and p53 gene polymorphisms modify selenium–ESCC relation.183 Studies showed that p53 Pro/Pro was related to ESCC risk as compared with p53 Arg/Arg homozygotes.183 Individuals with both the GSTP1 Ile/ Ile and p53 Pro/Pro genotypes have an increased risk of ESCC.183 Therefore, dietary selenium intake can alter an individual’s risk of developing EC. This risk can then be further increased if the polymorphisms in the GSTP1 and the p53 genes are also present.183
Conclusion
This review highlights the contribution that genetic polymorphisms make to the incidence of EC in a South African population, how these genetic polymorphisms are found in other population groups, especially Chinese populations, and the extent to which they contribute to the incidence of EC in these population groups. The characterization of these genetic polymorphisms may allow us to identify the molecules that can serve as lead drug targets as well as the new diagnostic and prognostic markers. These polymorphisms can best be studied through the use of large cohort studies that take into account the role played by environmental and lifestyle factors that may contribute to or protect an individual from EC. These studies would not only enable us to identify new genetic polymorphisms that are involved in EC, but also allow us to more accurately establish the role played by environmental hazards, such as drinking alcohol, smoking tobacco and consumption of Fusarium-contaminated maize. However, the future challenges include combining the results of these multiple studies in such a way so as to obtain a set of genetic signatures that can be used as population-specific prognostic or diagnostic markers, or even as targets for the development of new drugs. Furthermore, simultaneous analysis of multiple polymorphic genes would allow us to obtain a complete picture of their contribution in the development of EC.
Acknowledgment
We would like to thank the National Research Foundation and the Medical Research Council of South Africa for funding this research.
Disclosure
The authors report no conflicts of interest in this work.
References
Strickland NJ, Matsha T, Erasmus RT, Zaahl MG. Molecular analysis of ceruloplasmin in a South African cohort presenting with oesophageal cancer. Int J Cancer. 2012;131(3):623–632. | ||
World Health Organization. Global Status Report on Alcohol and Health. Geneva: World Health Organization; 2014. Available from: https://www.who.int/substance_abuse/publications/global_alcohol_report/en/. Accessed May 12, 2017. | ||
Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. | ||
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. | ||
Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381(9864):400–412. | ||
Hendricks D, Parker MI. Oesophageal cancer in Africa. IUBMB Life. 2002;53(4–5):263–268. | ||
Cooper SC, Day R, Brooks C, Livings C, Thomson CS, Trudgill NJ. The influence of deprivation and ethnicity on the incidence of esophageal cancer in England. Cancer Causes Control. 2009;20(8):1459–1467. | ||
Somdyala NI, Bradshaw D, Gelderblom WC, Parkin DM. Cancer incidence in a rural population of South Africa, 1998–2002. Int J Cancer. 2010;127(10):2420–2429. | ||
Kuwano H, Kato H, Miyazaki T, et al. Genetic alterations in esophageal cancer. Surg Today. 2005;35(1):7–18. | ||
Wu X, Chen VW, Ruiz B, Andrews P, Su LJ, Correa P. Incidence of esophageal and gastric carcinomas among American Asians/Pacific Islanders, whites, and blacks: subsite and histology differences. Cancer. 2006;106(3):683–692. | ||
van Blankenstein M, Looman CW, Hop WC, Bytzer P. The incidence of adenocarcinoma and squamous cell carcinoma of the esophagus: Barrett’s esophagus makes a difference. Am J Gastroenterol. 2005;100(4):766–774. | ||
Kollarova H, Machova L, Horakova D, Janoutova G, Janout V. Epidemiology of esophageal cancer – an overview article. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2007;151(1):17–28. | ||
Kamangar F, Diaw L, Wei WQ, et al. Serum pepsinogens and risk of esophageal squamous dysplasia. Int J Cancer. 2009;124(2):456–460. | ||
Cao B, Tian X, Li Y, et al. LMP7/TAP2 gene polymorphisms and HPV infection in esophageal carcinoma patients from a high incidence area in China. Carcinogenesis. 2005;26(7):1280–1284. | ||
Hyland PL, Freedman ND, Hu N, et al. Genetic variants in sex hormone metabolic pathway genes and risk of esophageal squamous cell carcinoma. Carcinogenesis. 2013;34(5):1062–1068. | ||
Segal I, Reinach SG, de Beer M. Factors associated with oesophageal cancer in Soweto, South Africa. Br J Cancer. 1988;58(5):681–686. | ||
Higginson J, Oettlé AG. Cancer incidence in the Bantu and “Cape Colored” races of South Africa: report of a cancer survey in the Transvaal (1953–55). J Natl Cancer Inst. 1960;24(3):589–671. | ||
Oettl AG. An epidemic of oesophageal carcinoma in Africa. S Afr Med J. 1963;37:435. | ||
Rose EF. Esophageal cancer in the Transkei: 1955–69. J Natl Cancer Inst. 1973;51(1):7–16. | ||
Jemal A, Bray F, Forman D, et al. Cancer burden in Africa and opportunities for prevention. Cancer. 2012;118(18):4372–4384. | ||
Blot WJ. Esophageal cancer trends and risk factors. Semin Oncol. 1994;21(4):403–410. | ||
Lagergren J, Bergström R, Lindgren A, Nyrén O. Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med. 1999;340(11):825–831. | ||
Sammon AM. Carcinogens and endemic squamous cancer of the oesophagus in Transkei, South Africa. Environmental initiation is the dominant factor; tobacco or other carcinogens of low potency or concentration are sufficient for carcinogenesis in the predisposed mucosa. Med Hypotheses. 2007;69(1):125–131. | ||
Makaula AN, Marasas WF, Venter FS, Badenhorst CJ, Bradshaw D, Swanevelder S. Oesophageal and other cancer patterns in four selected districts of the Transkei, southern Africa:1985–1990. Afr J Health Sci. 1996;3(1):11–15. | ||
Sumeruk R, Segal I, Te Winkel W, van der Merwe CF. Oesophageal cancer in three regions of South Africa. S Afr Med J. 1992;81(2):91–93. | ||
Dlamini Z, Bhoola K. Esophageal cancer in African blacks of Kwazulu natal, South Africa: an epidemiological brief. Ethn Dis. 2005;15(4):786–789. | ||
Chen M, Huang J, Zhu Z, Zhang J, Li K. Systematic review and meta-analysis of tumor biomarkers in predicting prognosis in esophageal cancer. BMC Cancer. 2013;13(1):539. | ||
Parkin DM, Stjernswärd J, Muir CS. Estimates of the worldwide frequency of twelve major cancers. Bull World Health Organ. 1984;62(2):163–182. | ||
Guo W, Blot WJ, Li JY, et al. A nested case–control study of oesophageal and stomach cancers in the Linxian Nutrition Intervention Trial. Int J Epidemiol. 1994;23(3):444–450. | ||
Lin Y, Totsuka Y, He Y, et al. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23(4):233–242. | ||
Wang AH, Sun CS, Li LS, Huang JY, Chen QS. Relationship of tobacco smoking CYP1A1 GSTM1 gene polymorphism and esophageal cancer in Xi’an. World J Gastroenterol. 2002;8(1):49–53. | ||
Long N, Moore MA, Chen W, et al. Cancer epidemiology and control in north-East Asia – past, present and future. Asian Pac J Cancer Prev. 2010;11 Suppl 2(Suppl 2):107–148. | ||
Barber JP. South Africa in The Twentieth Century: a Political History – in Search of a Nation State. Oxford, UK: Blackwell Publishers; 1999. | ||
Vaughan TL, Davis S, Kristal A, Thomas DB. Obesity, alcohol, and tobacco as risk factors for cancers of the esophagus and gastric cardia: adenocarcinoma versus squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 1995;4(2):85–92. | ||
Pacella-Norman R, Urban MI, Sitas F, et al. Risk factors for oesophageal, lung, oral and laryngeal cancers in black South Africans. Br J Cancer. 2002;86(11):1751–1756. | ||
An Y, Jin G, Wang H, et al. Polymorphisms in hMLH1 and risk of early-onset lung cancer in a southeast Chinese population. Lung Cancer. 2008;59(2):164–170. | ||
Lin Y, Totsuka Y, Shan B, et al. Esophageal cancer in high-risk areas of China: research progress and challenges. Ann Epidemiol. 2017;27(3):215–221. | ||
Schandl L, Malfertheiner P, Ebert MP. Prevention of gastric cancer by Helicobacter pylori eradication? Results from clinical intervention studies. Dig Dis. 2002;20(1):18–22. | ||
Sewram V, Sitas F, O’Connell D, Myers J. Tobacco and alcohol as risk factors for oesophageal cancer in a high incidence area in South Africa. Cancer Epidemiol. 2016;41:113–121. | ||
Reddy P, Zuma K, Shisana O, Kim J, Sewpaul R. Prevalence of tobacco use among adults in South Africa: results from the first South African National Health and Nutrition Examination Survey. S Afr Med J. 2015;105(8):648–655. | ||
Zenzes MT. Smoking and reproduction: gene damage to human gametes and embryos. Hum Reprod Update. 2000;6(2):122–131. | ||
Li Q, Hsia J, Yang G. Prevalence of smoking in China in 2010. N Engl J Med. 2011;364(25):2469–2470. | ||
Wang LD, Zhou FY, Li XM, et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42(9):759–763. | ||
Liang H, Wang J, Xiao H, et al. Estimation of cancer incidence and mortality attributable to alcohol drinking in China. BMC Public Health. 2010;10(1):730. | ||
Sewram V, Sitas F, O’Connell D, Myers J. Diet and esophageal cancer risk in the Eastern Cape Province of South Africa. Nutr Cancer. 2014;66(5):791–799. | ||
Jaskiewicz K. Oesophageal carcinoma: cytopathology and nutritional aspects in aetiology. Anticancer Res. 1989;9(6):1847–1852. | ||
Taylor PR, Li B, Dawsey SM, et al. Prevention of esophageal cancer: the nutrition intervention trials in Linxian, China. Linxian Nutrition Intervention Trials Study Group. Cancer Res. 1994;54(7 Suppl):2029s–2031. | ||
Isaacson C. The change of the staple diet of black South Africans from sorghum to maize (corn) is the cause of the epidemic of squamous carcinoma of the oesophagus. Med Hypotheses. 2005;64(3):658–660. | ||
Pillay V, Isaacson C, Mothobi P, et al. Carcinogenic nitrosamines in traditional beer as the cause of oesophageal squamous cell carcinoma in black South Africans. S Afr Med J. 2015;105(8):656–658. | ||
Pink RC, Bailey TA, Iputo JE, Sammon AM, Woodman AC, Carter DR. Molecular basis for maize as a risk factor for esophageal cancer in a South African population via a prostaglandin E2 positive feedback mechanism. Nutr Cancer. 2011;63(5):714–721. | ||
Sammon AM, Morgan A. Dietary fat and salivary prostaglandin E2. Prostaglandins Other Lipid Mediat. 2002;67(2):137–141. | ||
Sammon AM, Mguni M, Mapele L, Awotedu KO, Iputo JE. Bimodal distribution of fasting gastric acidity in a rural African population. S Afr Med J. 2003;93(10):786–788. | ||
Sammon AM. Protease inhibitors and carcinoma of the esophagus. Cancer. 1998;83(3):405–408. | ||
Morgan G. Deleterious effects of prostaglandin E2 in oesophageal carcinogenesis. Med Hypotheses. 1997;48(2):177–181. | ||
Gupta RA, Tejada LV, Tong BJ, et al. Cyclooxygenase-1 is overexpressed and promotes angiogenic growth factor production in ovarian cancer. Cancer Res. 2003;63(5):906–911. | ||
Akbari MR, Malekzadeh R, Shakeri R, et al. Candidate gene association study of esophageal squamous cell carcinoma in a high-risk region in Iran. Cancer Res. 2009;69(20):7994–8000. | ||
Yu L, Wu WK, Li ZJ, Li HT, Wu YC, Cho CH. Prostaglandin E(2) promotes cell proliferation via protein kinase C/extracellular signal regulated kinase pathway-dependent induction of c-Myc expression in human esophageal squamous cell carcinoma cells. Int J Cancer. 2009;125(11):2540–2546. | ||
Yang CS. Research on esophageal cancer in China: a review. Cancer Res. 1980;40(8 Pt 1):2633–2644. | ||
Kgomo M, Elnagar AA, Mokoena T, Jeske C, Nagel GJ. Prevalence of Helicobacter pylori infection in patients with squamous cell carcinoma of the oesophagus. A descriptive case series study. J Gastrointest Cancer. 2016;47(4):396–398. | ||
Kamangar F, Qiao YL, Blaser MJ, et al. Helicobacter pylori and oesophageal and gastric cancers in a prospective study in China. Br J Cancer. 2007;96(1):172–176. | ||
Schäfer G, Kabanda S, van Rooyen B, Marušič MB, Banks L, Parker MI. The role of inflammation in HPV infection of the oesophagus. BMC Cancer. 2013;13(1):185. | ||
Koshiol J, Wei WQ, Kreimer AR, et al. No role for human papillomavirus in esophageal squamous cell carcinoma in China. Int J Cancer. 2010;127(1):93–100. | ||
Wu M, Liu AM, Kampman E, et al. Green tea drinking, high tea temperature and esophageal cancer in high- and low-risk areas of Jiangsu Province, China: a population-based case–control study. Int J Cancer. 2009;124(8):1907–1913. | ||
Willem P, Brown J, Schouten J. A novel approach to simultaneously scan genes at fragile sites. BMC Cancer. 2006;6(1):205. | ||
Stein CK, Glover TW, Palmer JL, Glisson BS. Direct correlation between FRA3B expression and cigarette smoking. Genes Chromosomes Cancer. 2002;34(3):333–340. | ||
Thorland EC, Myers SL, Gostout BS, Smith DI. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene. 2003;22(8):1225–1237. | ||
Capaccio P, Ottaviani F, Cuccarini V, Cenzuales S, Cesana BM, Pignataro L. Association between methylenetetrahydrofolate reductase polymorphisms, alcohol intake and oropharyngolaryngeal carcinoma in northern Italy. J Laryngol Otol. 2005;119(5):371–376. | ||
Yokoyama T, Saito K, Lwin H, et al. Epidemiological evidence that acetaldehyde plays a significant role in the development of decreased serum folate concentration and elevated mean corpuscular volume in alcohol drinkers. Alcohol Clin Exp Res. 2005;29(4):622–630. | ||
Knasmüller S, Bresgen N, Kassie F, et al. Genotoxic effects of three Fusarium mycotoxins, fumonisin B1, moniliformin and vomitoxin in bacteria and in primary cultures of rat hepatocytes. Mutat Res. 1997;391(1–2):39–48. | ||
Odera JO, Odera E, Githang’a J, et al. Esophageal cancer in Kenya. Am J Dig Dis (Madison). 2017;4(3):23–33. | ||
Cheung WY, Liu G. Genetic variations in esophageal cancer risk and prognosis. Gastroenterol Clin North Am. 2009;38(1):75–91. | ||
Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and esophageal cancer risk. Int J Cancer. 2007;121(8):1643–1658. | ||
Vogelsang M, Wang Y, Veber N, Mwapagha LM, Parker MI. The cumulative effects of polymorphisms in the DNA mismatch repair genes and tobacco smoking in oesophageal cancer risk. PLoS One. 2012;7(5):e36962. | ||
Cargill M, Altshuler D, Ireland J, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22(3):231–238. | ||
Singh MS, Michael M. Role of xenobiotic metabolic enzymes in cancer epidemiology. Methods Mol Biol. 2009;472:243–264. | ||
Liska DJ. The detoxification enzyme systems. Altern Med Rev. 1998;3(3):187–198. | ||
Wang LD, Zheng S, Liu B, Zhou JX, Li YJ, Li JX. CYP1A1, GSTs and mEH polymorphisms and susceptibility to esophageal carcinoma: study of population from a high-incidence area in North China. World J Gastroenterol. 2003;9(7):1394–1397. | ||
Abnet CC, Freedman ND, Hu N, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42(9):764–767. | ||
Lao-Sirieix P, Caldas C, Fitzgerald RC. Genetic predisposition to gastro-oesophageal cancer. Curr Opin Genet Dev. 2010;20(3):210–217. | ||
Nebert DW, Roe AL, Vandale SE, Bingham E, Oakley GG. NAD (P) H: quinone oxidoreductase (NQO1) polymorphism, exposure to benzene, and predisposition to disease: a huge review. Genet Med. 2002;4(2):62–70. | ||
Kuehl BL, Paterson JW, Peacock JW, Paterson MC, Rauth AM. Presence of a heterozygous substitution and its relationship to DT-diaphorase activity. Br J Cancer. 1995;72(3):555–561. | ||
Klaidman LK, Leung AC, Adams JD. High-performance liquid chromatography analysis of oxidized and reduced pyridine dinucleotides in specific brain regions. Anal Biochem. 1995;228(2):312–317. | ||
Radjendirane V, Joseph P, Lee YH, et al. Disruption of the DT diaphorase (NQO1) gene in mice leads to increased menadione toxicity. J Biol Chem. 1998;273(13):7382–7389. | ||
Long DJ, Gaikwad A, Multani A, et al. Disruption of the NAD(P)H:quinone oxidoreductase 1 (NQO1) gene in mice causes myelogenous hyperplasia. Cancer Res. 2002;62(11):3030–3036. | ||
Yanling H, Yuhong Z, Wenwu H, Lei X, Mingwu C. NQO1 C609T polymorphism and esophageal cancer risk: a huge review and meta-analysis. BMC Med Genet. 2013;14(1):31. | ||
Salaspuro M. Acetaldehyde and gastric cancer. J Dig Dis. 2011;12(2):51–59. | ||
Yang SJ, Wang HY, Li XQ, et al. Genetic polymorphisms of ADH2 and ALDH2 association with esophageal cancer risk in southwest China. World J Gastroenterol. 2007;13(43):5760–5764. | ||
Li QD, Li H, Wang MS, et al. Multi-susceptibility genes associated with the risk of the development stages of esophageal squamous cell cancer in Feicheng County. BMC Gastroenterol. 2011;11:74. | ||
Li DP, Dandara C, Walther G, Parker MI. Genetic polymorphisms of alcohol metabolising enzymes: their role in susceptibility to oesophageal cancer. Clin Chem Lab Med. 2008;46(3):323–328. | ||
Bye H, Prescott NJ, Matejcic M, et al. Population-specific genetic associations with oesophageal squamous cell carcinoma in South Africa. Carcinogenesis. 2011;32(12):1855–1861. | ||
Li H, Borinskaya S, Yoshimura K, et al. Refined geographic distribution of the oriental ALDH2*504Lys (nee 487Lys) variant. Ann Hum Genet. 2009;73(Pt 3):335–345. | ||
Wilding G. The importance of steroid hormones in prostate cancer. Cancer Surv. 1992;14:113–130. | ||
Ferro P, Catalano MG, Raineri M, et al. Somatic alterations of the androgen receptor CAG repeat in human colon cancer delineate a novel mutation pathway independent of microsatellite instability. Cancer Genet Cytogenet. 2000;123(1):35–40. | ||
Hackenberg R, Schulz KD. Androgen receptor mediated growth control of breast cancer and endometrial cancer modulated by antiandrogen- and androgen-like steroids. J Steroid Biochem Mol Biol. 1996;56(1–6 Spec No):113–117. | ||
Nagasue N, Yu L, Yukaya H, Kohno H, Nakamura T. Androgen and oestrogen receptors in hepatocellular carcinoma and surrounding liver parenchyma: impact on intrahepatic recurrence after hepatic resection. Br J Surg. 1995;82(4):542–547. | ||
Yamashita Y, Hirai T, Mukaida H, et al. Detection of androgen receptors in human esophageal cancer. Jpn J Surg. 1989;19(2):195–202. | ||
Lubahn D, Joseph D, Sullivan P, Willard H, French F, Wilson E. Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science. 1988;240(4850):327–330. | ||
Quigley CA, De Bellis A, Marschke KB, El-Awady MK, Wilson EM, French FS. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16(3):271–321. | ||
Dehm SM, Tindall DJ. Androgen receptor structural and functional elements: role and regulation in prostate cancer. Mol Endocrinol. 2007;21(12):2855–2863. | ||
Claessens F, Denayer S, Van Tilborgh N, Kerkhofs S, Helsen C, Haelens A. Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl Recept Signal. 2008;6:e008. | ||
Edwards A, Hammond HA, Jin L, Caskey CT, Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992;12(2):241–253. | ||
Jiang W, Zhang YJ, Kahn SM, et al. Altered expression of the cyclin D1 and retinoblastoma genes in human esophageal cancer. Proc Natl Acad Sci U S A. 1993;90(19):9026–9030. | ||
Yin J, Sang Y, Zheng L, et al. Uracil-DNA glycosylase (UNG) rs246079 G/A polymorphism is associated with decreased risk of esophageal cancer in a Chinese population. Med Oncol. 2014;31(11):272. | ||
Du Plessis L, Dietzsch E, Van Gele M, et al. Mapping of novel regions of DNA gain and loss by comparative genomic hybridization in esophageal carcinoma in the black and colored populations of South Africa. Cancer Res. 1999;59(8):1877–1883. | ||
Hu N, Roth MJ, Polymeropolous M, et al. Identification of novel regions of allelic loss from a genomewide scan of esophageal squamous-cell carcinoma in a high-risk Chinese population. Genes Chromosomes Cancer. 2000;27(3):217–228. | ||
Sleddens HF, Oostra BA, Brinkmann AO, Trapman J. Trinucleotide (GGN) repeat polymorphism in the human androgen receptor (AR) gene. Hum Mol Genet. 1993;2(4):493. | ||
Hsing AW, Gao YT, Wu G, et al. Polymorphic CAG and Ggn repeat lengths in the androgen receptor gene and prostate cancer risk: a population-based case–control study in China. Cancer Res. 2000;60(18):5111–5116. | ||
Dietzsch E, Laubscher R, Parker MI. Esophageal cancer risk in relation to GGC and CAG trinucleotide repeat lengths in the androgen receptor gene. Int J Cancer. 2003;107(1):38–45. | ||
Ferro P, Catalano MG, Dell’Eva R, Fortunati N, Pfeffer U. The androgen receptor CAG repeat: a modifier of carcinogenesis? Mol Cell Endocrinol. 2002;193(1–2):109–120. | ||
Marchewka Z, Piwowar A, Ruzik S, Długosz A. Glutathione S-transferases class Pi and Mi and their significance in oncology. Postepy Hig Med Dosw (Online). 2017;71(0):541–550. | ||
Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. | ||
Nelson HH, Wiencke JK, Christiani DC, et al. Ethnic differences in the prevalence of the homozygous deleted genotype of glutathione S-transferase theta. Carcinogenesis. 1995;16(5):1243–1246. | ||
Fryer AA. The glutathione S-transferases: influence of polymorphism on cancer susceptibility. IARC Sci Publ. 1999;148:231–249. | ||
Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30(6):445–520. | ||
Dutton MF, Fumonisins DMF. Fumonisins, mycotoxins of increasing importance: their nature and their effects. Pharmacol Ther. 1996;70(2):137–161. | ||
Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149(7):1700–1715. | ||
Pearson WR, Vorachek WR, Xu SJ, Berger R, Hart I, Vannais D, Patterson D. Identification of class-mu glutathione transferase genes GSTM1–GSTM5 on human chromosome 1p13. Am J Hum Genet. 1993;53(1):220–233. | ||
Coggan M, Whitbread L, Whittington A, Board P. Structure and organization of the human theta-class glutathione S-transferase and d-dopachrome tautomerase gene complex. Biochem J. 1998;334(Pt 3):617–623. | ||
He B, Pan Y, Cho WC, et al. The association between four genetic variants in microRNAs (rs11614913, rs2910164, rs3746444, rs2292832) and cancer risk: evidence from published studies. PLoS One. 2012;7(11): e49032. | ||
Ge H, Cao YY, Chen LQ, et al. PTEN polymorphisms and the risk of esophageal carcinoma and gastric cardiac carcinoma in a high incidence region of China. Dis Esophagus. 2008;21(5):409–415. | ||
Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143(1):35–47. | ||
Ogawa R, Ishiguro H, Kuwabara Y, et al. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Med Mol Morphol. 2009;42(2):102–109. | ||
Yao L, Zhang Y, Zhu Q, et al. Downregulation of microRNA-1 in esophageal squamous cell carcinoma correlates with an advanced clinical stage and its overexpression inhibits cell migration and invasion. Int J Mol Med. 2015;35(4):1033–1041. | ||
Liu R, Gu J, Jiang P, et al. DNMT1–microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res. 2015;21(4):854–863. | ||
Suzuki H, Maruyama R, Yamamoto E, Kai M. DNA methylation and microRNA dysregulation in cancer. Mol Oncol. 2012;6(6):567–578. | ||
Burger HM, Lombard MJ, Shephard GS, Rheeder JR, van der Westhuizen L, Gelderblom WC. Dietary fumonisin exposure in a rural population of South Africa. Food Chem Toxicol. 2010;48(8–9):2103–2108. | ||
Guled M, Lahti L, Lindholm PM, et al. CDKN2A, NF2, and Jun are dysregulated among other genes by miRNAs in malignant mesothelioma – a miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48(7):615–623. | ||
Wang Y, Vogelsang M, Schäfer G, Matejcic M, Parker MI. MicroRNA polymorphisms and environmental smoke exposure as risk factors for oesophageal squamous cell carcinoma. PLoS One. 2013;8(10):e78520. | ||
Lin J, Huang S, Wu S, et al. MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis. 2011;32(11):1641–1647. | ||
Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila). 2008;1(6):460–469. | ||
Guo H, Wang K, Xiong G, et al. A functional variant in microRNA-146a is associated with risk of esophageal squamous cell carcinoma in Chinese Han. Fam Cancer. 2010;9(4):599–603. | ||
Wei J, Zheng L, Liu S, et al. miR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese population. Hum Immunol. 2013;74(9):1199–1205. | ||
Yin J, Wang X, Zheng L, et al. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A polymorphisms are associated with the risk of esophageal cancer in a Chinese population. PLoS One. 2013;8(11):e80570. | ||
Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68(7):2530–2537. | ||
Wei Q, Eicher SA, Guan Y, et al. Reduced expression of hMLH1 and hGTBP/hMSH6: a risk factor for head and neck cancer. Cancer Epidemiol Biomarkers Prev. 1998;7(4):309–314. | ||
Hirao T, Nelson HH, Ashok TD, et al. Tobacco smoke-induced DNA damage and an early age of smoking initiation induce chromosome loss at 3p21 in lung cancer. Cancer Res. 2001;61(2):612–615. | ||
Bavi PP, Bu R, Uddin S, Al-Kuraya KS. MMP7 polymorphisms – a new tool in molecular pathology to understand esophageal cancer. Saudi J Gastroenterol. 2011;17(5):299–300. | ||
Lin DX, Tang YM, Peng Q, Lu SX, Ambrosone CB, Kadlubar FF. Susceptibility to esophageal cancer and genetic polymorphisms in glutathione S-transferases T1, P1, and M1 and cytochrome P450 2E1. Cancer Epidemiol Biomarkers Prev. 1998;7(11):1013–1018. | ||
Schrader CE, Vardo J, Stavnezer J. Role for mismatch repair proteins MSH2, MLH1, and pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J Exp Med. 2002;195(3):367–373. | ||
Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(6):2073–2087.e. | ||
Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015;16(7):30. | ||
Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. | ||
Naidoo R, Ramburan A, Reddi A, Chetty R. Aberrations in the mismatch repair genes and the clinical impact on oesophageal squamous carcinomas from a high incidence area in South Africa. J Clin Pathol. 2005;58(3):281–284. | ||
Uehara H, Miyamoto M, Kato K, et al. Deficiency of hMLH1 and hMSH2 expression is a poor prognostic factor in esophageal squamous cell carcinoma. J Surg Oncol. 2005;92(2):109–115. | ||
Vogelsang M, Paccez JD, Schäfer G, Dzobo K, Zerbini LF, Parker MI. Aberrant methylation of the MSH3 promoter and distal enhancer in esophageal cancer patients exposed to first-hand tobacco smoke. J Cancer Res Clin Oncol. 2014;140(11):1825–1833. | ||
Campregher C, Schmid G, Ferk F, et al. MSH3-deficiency initiates EMAST without oncogenic transformation of human colon epithelial cells. PLoS One. 2012;7(11):e50541. | ||
Park JM, Huang S, Tougeron D, Sinicrope FA. MSH3 mismatch repair protein regulates sensitivity to cytotoxic drugs and a histone deacetylase inhibitor in human colon carcinoma cells. PLoS One. 2013;8(5):e65369. | ||
Takahashi M, Koi M, Balaguer F, Boland CR, Goel A. MSH3 mediates sensitization of colorectal cancer cells to cisplatin, oxaliplatin, and a poly(ADP-ribose) polymerase inhibitor. J Biol Chem. 2011;286(14):12157–12165. | ||
Huang J, Okuka M, Lu W, et al. Telomere shortening and DNA damage of embryonic stem cells induced by cigarette smoke. Reprod Toxicol. 2013;35:89–95. | ||
Ling ZQ, Li P, Ge MH, Hu FJ, Fang XH, Dong ZM, Mao WM. Aberrant methylation of different DNA repair genes demonstrates distinct prognostic value for esophageal cancer. Dig Dis Sci. 2011;56(10):2992–3004. | ||
Wu D, Chen X, Xu Y, et al. Prognostic value of MLH1 promoter methylation in male patients with esophageal squamous cell carcinoma. Oncol Lett. 2017;13(4):2745–2750. | ||
Guengerich FP. Cytochromes P450, drugs, and diseases. Mol Interv. 2003;3(4):194–204. | ||
Perera FP. Environment and cancer: who are susceptible? Science. 1997;278(5340):1068–1073. | ||
Strassburg CP, Strassburg A, Nguyen N, Li Q, Manns MP, Tukey RH. Regulation and function of family 1 and family 2 UDP-glucuronosyltransferase genes (UGT1A, UGT2B) in human oesophagus. Biochem J. 1999;382(Pt 2):489–498. | ||
Adams CH, Werely CJ, Victor TC, Hoal EG, Rossouw G, van Helden PD. Allele frequencies for glutathione S-transferase and N-acetyltransferase 2 differ in African population groups and may be associated with oesophageal cancer or tuberculosis incidence. Clin Chem Lab Med. 2003;41(4):600–605. | ||
Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–173. | ||
Umeno M, McBride OW, Yang CS, Gelboin HV, Gonzalez FJ. Human ethanol-inducible P450IIE1: complete gene sequence, promoter characterization, chromosome mapping, and cDNA-directed expression. Biochemistry. 1988;27(25):9006–9013. | ||
Dandara C, Ballo R, Parker MI. CYP3A5 genotypes and risk of oesophageal cancer in two South African populations. Cancer Lett. 2005;225(2):275–282. | ||
Gellner K, Eiselt R, Hustert E, et al. Genomic organization of the human CYP3A locus: identification of a new, inducible CYP3A gene. Pharmacogenetics. 2001;11(2):111–121. | ||
Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54(10):1271–1294. | ||
Roy JN, Lajoie J, Zijenah LS, et al. CYP3A5 genetic polymorphisms in different ethnic populations. Drug Metab Dispos. 2005;33(7):884–887. | ||
Lechevrel M, Casson AG, Wolf CR, Hardie LJ, Flinterman MB, Montesano R, Wild CP. Characterization of cytochrome P450 expression in human oesophageal mucosa. Carcinogenesis. 1999;20(2):243–248. | ||
Marasas WF. Discovery and occurrence of the fumonisins: a historical perspective. Environ Health Perspect. 2001;109(Suppl 2):239–243. | ||
Spotti M, Maas RF, de Nijs CM, Fink-Gremmels J. Effect of fumonisin B(1) on rat hepatic P450 system. Environ Toxicol Pharmacol. 2000;8(3):197–204. | ||
Löfdahl HE, Lu Y, Lagergren J. Sex-specific risk factor profile in oesophageal adenocarcinoma. Br J Cancer. 2008;99(9):1506–1510. | ||
Bodelon C, Anderson GL, Rossing MA, Chlebowski RT, Ochs-Balcom HM, Vaughan TL. Hormonal factors and risks of esophageal squamous cell carcinoma and adenocarcinoma in postmenopausal women. Cancer Prev Res (Phila). 2011;4(6):840–850. | ||
Gallus S, Bosetti C, Franceschi S, et al. Oesophageal cancer in women: tobacco, alcohol, nutritional and hormonal factors. Br J Cancer. 2001;85(3):341–345. | ||
Wang QM, Qi YJ, Jiang Q, et al. Relevance of serum estradiol and estrogen receptor beta expression from a high-incidence area for esophageal squamous cell carcinoma in China. Med Oncol. 2011;28(1):188–193. | ||
Tihan T, Harmon JW, Wan X, et al. Evidence of androgen receptor expression in squamous and adenocarcinoma of the esophagus. Anticancer Res. 2001;21(4b):3107–3114. | ||
Hanna IH, Dawling S, Roodi N, Guengerich FP, Parl FF. Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res. 2000;60(13):3440–3444. | ||
Ryan-Harshman M, Aldoori W. The relevance of selenium to immunity, cancer, and infectious/inflammatory diseases. Can J Diet Pract Res. 2005;66(2):98–102. | ||
Kocdor H, Cehreli R, Kocdor MA, et al. Toxicity induced by the chemical carcinogen 7,12-dimethylbenz[a]anthracene and the protective effects of selenium in Wistar rats. J Toxicol Environ Health A. 2005;68(9):693–701. | ||
Longtin R. Selenium for prevention: eating your way to better DNA repair? J Natl Cancer Inst. 2003;95(2):98–100. | ||
Zhou N, Xiao H, Li TK, et al. DNA damage-mediated apoptosis induced by selenium compounds. J Biol Chem. 2003;278(32):29532–29537. | ||
Smith ML, Lancia JK, Mercer TI, Ip C. Selenium compounds regulate p53 by common and distinctive mechanisms. Anticancer Res. 2004;24(3a):1401–1408. | ||
Seo YR, Kelley MR, Smith ML. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc Natl Acad Sci U S A. 2002;99(22):14548–14553. | ||
Ali-Osman F, Akande O, Antoun G, Mao JX, Buolamwini J. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem. 1997;272(15):10004–10012. | ||
Bennett WP, Hollstein MC, Metcalf RA, et al. P53 mutation and protein accumulation during multistage human esophageal carcinogenesis. Cancer Res. 1992;52(21):6092–6097. | ||
Lee JM, Lee YC, Yang SY, et al. Genetic polymorphisms of p53 and GSTP1, but not NAT2, are associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 2000;89(5):458–464. | ||
Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7(2):961–963. | ||
Lu XM, Zhang YM, Lin RY, et al. P53 polymorphism in human papillomavirus-associated Kazakh’s esophageal cancer in Xinjiang, China. World J Gastroenterol. 2004;10(19):2775–2778. | ||
Li X, Dumont P, della Pietra A, Shetler C, Murphy ME. The codon 47 polymorphism in p53 is functionally significant. J Biol Chem. 2005;280(25):24245–24251. | ||
Cai L, Mu LN, Lu H, et al. Dietary selenium intake and genetic polymorphisms of the GSTP1 and p53 genes on the risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15(2):294–300. | ||
Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30(1):1–12. | ||
South African National Cancer Registry. Cancer in South Africa; 2014. Available from: www.ncr.ac.za. Accessed October 15, 2018. | ||
World Health Organization. Prevalence of Tobacco Smoking. Geneva: World Health Organization; 2015. Available from: www.who.int/gho/tobacco/use/en/. Accessed August 12, 2017. | ||
World Health Organization. Global Status Report on Alcohol and Health. Geneva: World Health Organization; 2014. Available from: https://www.who.int/gho/tobacco/use/en/. Accessed May 12, 2017. | ||
Torres-Aguilera M, Remes Troche JM. Achalasia and esophageal cancer: risks and links. Clin Exp Gastroenterol. 2018;11:309–316. | ||
Coleman HG, Xie SH, Lagergren J. The epidemiology of esophageal adenocarcinoma. Gastroenterology. 2018;154(2):390–405. | ||
Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154(2):360–373. | ||
Singh S, Devanna S, Edakkanambeth Varayil J, Murad MH, Iyer PG. Physical activity is associated with reduced risk of esophageal cancer, particularly esophageal adenocarcinoma: a systematic review and meta-analysis. BMC Gastroenterol. 2014;14:101. | ||
Erőss B, Farkas N, Vincze Á, et al. Helicobacter pylori infection reduces the risk of Barrett’s esophagus: a meta-analysis and systematic review. Helicobacter. 2018;23(4):e12504. | ||
Xie FJ, Zhang YP, Zheng QQ, et al. Helicobacter pylori infection and esophageal cancer risk: an updated meta-analysis. World J Gastroenterol. 2013;19(36):6098–6107. | ||
Brusselaers N, Maret-Ouda J, Konings P, El-Serag HB, Lagergren J. Menopausal hormone therapy and the risk of esophageal and gastric cancer. Int J Cancer. 2017;140(7):1693–1699. | ||
Li P, Cheng R, Zhang S. Aspirin and esophageal squamous cell carcinoma: bedside to bench. Chin Med J (Engl). 2014;127(7):1365–1369. | ||
Qu Y, Zhang S, Cui L, et al. Two novel polymorphisms in PLCE1 are associated with the susceptibility to esophageal squamous cell carcinoma in Chinese population. Dis Esophagus. 2017;30(1):1–7. | ||
Hu H, Yang J, Sun Y, et al. Putatively functional PLCE1 variants and susceptibility to esophageal squamous cell carcinoma (EscC): a case–control study in eastern Chinese populations. Ann Surg Oncol. 2012;19(7):2403–2410. | ||
Bye H, Prescott NJ, Lewis CM, et al. Distinct genetic association at the PLCE1 locus with oesophageal squamous cell carcinoma in the South African population. Carcinogenesis. 2012;33(11):2155–2161. | ||
Wang L, Tang W, Chen S, et al. N-acetyltransferase 2 polymorphisms and risk of esophageal cancer in a Chinese population. PLoS One. 2014;9(2):e87783. | ||
Matejcic M, Vogelsang M, Wang Y, Iqbal Parker M, Parker IM. NAT1 and NAT2 genetic polymorphisms and environmental exposure as risk factors for oesophageal squamous cell carcinoma: a case–control study. BMC Cancer. 2015;15:150. | ||
Abnet CC, Wang Z, Song X, et al. Genotypic variants at 2q33 and risk of esophageal squamous cell carcinoma in China: a meta-analysis of genome-wide association studies. Hum Mol Genet. 2012;21(9):2132–2141. | ||
Hao XD, Yang Y, Song X, et al. Correlation of telomere length shortening with TP53 somatic mutations, polymorphisms and allelic loss in breast tumors and esophageal cancer. Oncol Rep. 2013;29(1):226–236. | ||
Zhou L, Yuan Q, Yang M. A functional germline variant in the p53 polyadenylation signal and risk of esophageal squamous cell carcinoma. Gene. 2012;506(2):295–297. | ||
Vos M, Adams CH, Victor TC, van Helden PD. Polymorphisms and mutations found in the regions flanking exons 5 to 8 of the TP53 gene in a population at high risk for esophageal cancer in South Africa. Cancer Genet Cytogenet. 2003;140(1):23–30. | ||
Gao Y, He Y, Xu J, et al. Genetic variants at 4q21, 4q23 and 12q24 are associated with esophageal squamous cell carcinoma risk in a Chinese population. Hum Genet. 2013;132(6):649–656. | ||
Liu P, Zhao HR, Li F, et al. Correlations of ALDH2 rs671 and C12orf30 rs4767364 polymorphisms with increased risk and prognosis of esophageal squamous cell carcinoma in the Kazak and Han populations in Xinjiang Province. J Clin Lab Anal. 2018;32(2): e22248. | ||
Guo YM, Wang Q, Liu YZ, Chen HM, Qi Z, Guo QH. Genetic polymorphisms in cytochrome P4502E1, alcohol and aldehyde dehydrogenases and the risk of esophageal squamous cell carcinoma in Gansu Chinese males. World J Gastroenterol. 2008;14(9):1444–1449. | ||
Townsend DM, Tew KD, Tapiero H. The importance of glutathione in human disease. Biomed Pharmacother. 2003;57(3–4):145–155. | ||
Li D, Dandara C, Parker MI. The 341C/T polymorphism in the GSTP1 gene is associated with increased risk of oesophageal cancer. BMC Genet. 2010;11(1):47. | ||
Matejcic M, Li D, Prescott NJ, Lewis CM, Mathew CG, Parker MI. Association of a deletion of GSTT2B with an altered risk of oesophageal squamous cell carcinoma in a South African population: a case–control study. PLoS One. 2011;6(12):e29366. | ||
Gao CM, Takezaki T, Wu JZ, et al. Glutathione-S-transferases M1 (GSTM1) and GSTT1 genotype, smoking, consumption of alcohol and tea and risk of esophageal and stomach cancers: a case–control study of a high-incidence area in Jiangsu Province, China. Cancer Lett. 2002;188(1–2):95–102. | ||
Dandara C, Li DP, Walther G, Parker MI. Gene–environment interaction: the role of SULT1A1 and CYP3A5 polymorphisms as risk modifiers for squamous cell carcinoma of the oesophagus. Carcinogenesis. 2006;27(4):791–797. | ||
Bolt HM, Thier R. Relevance of the deletion polymorphisms of the glutathione S-transferases GSTT1 and GSTM1 in pharmacology and toxicology. Curr Drug Metab. 2006;7(6):613–628. | ||
Zhao Y, Marotta M, Eichler EE, Eng C, Tanaka H. Linkage disequilibrium between two high-frequency deletion polymorphisms: implications for association studies involving the glutathione-S transferase (GST) genes. PLoS Genet. 2009;5(5):e1000472. | ||
Li T, Suo Q, He D, et al. Esophageal cancer risk is associated with polymorphisms of DNA repair genes MSH2 and WRN in Chinese population. J Thorac Oncol. 2012;7(2):448–452. | ||
Hayashi S, Watanabe J, Kawajiri K. Genetic polymorphisms in the 5’-flanking region change transcriptional regulation of the human cytochrome P450IIE1 gene. J Biochem. 1991;110(4):559–565. | ||
Lu XM, Zhang YM, Lin RY, et al. Relationship between genetic polymorphisms of metabolizing enzymes CYP2E1, GSTM1 and Kazakh’s esophageal squamous cell cancer in Xinjiang, China. World J Gastroenterol. 2005;11(24):3651–3654. | ||
Zheng H, Zhao Y. Association of CYP1A1 MspI polymorphism in the esophageal cancer risk: a meta-analysis in the Chinese population. Eur J Med Res. 2015;20(1):46. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





