Back to Journals » Infection and Drug Resistance » Volume 12
Escherichia coli belonging to ST131 rarely transfers blactx-m-15 to fecal Escherichia coli
Authors Thingholm KR, Hertz FB , Løbner-Olesen A, Frimodt-Møller N, Nielsen KL
Received 13 March 2019
Accepted for publication 23 May 2019
Published 6 August 2019 Volume 2019:12 Pages 2429—2435
DOI https://doi.org/10.2147/IDR.S208536
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Karen Rønø Thingholm,1 Frederik Boëtius Hertz,1,2 Anders Løbner-Olesen,3 Niels Frimodt-Møller,1 Karen Leth Nielsen1
1Department of Clinical Microbiology, Rigshospitalet, Copenhagen 2100, Denmark; 2Department of Clinical Microbiology, Herlev and Gentofte Hospital, Herlev 2730, Denmark; 3Department of Biology, Section for Functional Genomics and Center for Bacterial Stress Response and Persistence, University of Copenhagen, Copenhagen, Denmark
Background: Extended spectrum beta-lactamase (ESBL)-producing Escherichia coli (E. coli) causing urinary tract infections often belong to sequence type 131 (ST131), serotype O25, carrying blaCTX-M-15.
Aim: The main aim of this study was to examine the conjugational frequencies of E. coli with plasmids carrying blaCTX-M-15 to E. coli isolates from the fecal flora of healthy humans to determine whether ST131 is more likely to uptake or donate ESBL resistance compared to other E. coli clones.
Methods: Donors and recipients were all clinical isolates and did not harbor plasmids with identical incompatibility groups (Inc-groups) based on in silico analyses of Inc-groups and restriction/modification systems (R/M-systems). The in vitro conjugation experiments were performed as filter conjugation with verification of transconjugants by random amplified polymorphic DNA (RAPD) PCR and blaCTX-M-15 PCR.
Results: The frequencies of conjugation with blaCTX-M-15-carrying plasmids were found to be very rare with detectable conjugation frequencies in the range of 4x10−9–7x10−7 transconjugants/recipient. Recipients of O25/ST131 type yielded significantly lower conjugation frequencies compared to recipients of other O-types (P=0.004). The applied ST131/O25 donors did not yield detectable levels of transconjugants regardless of the applied recipient. Presence of sub-MIC levels of ampicillin increased plasmid transfer frequencies x100 fold (P=0.07).
Conclusion: The results indicate that blaCTX-M-15 is rarely transferred by conjugation to E. coli isolates of the intestinal flora, even when the gene is plasmid-borne.
Keywords: horizontal gene transfer, antimicrobial resistance, Escherichia coli, molecular typing, ST131, faecal isolates
Introduction
Escherichia coli is a part of the normal intestinal flora of healthy individuals with increasing levels of extended-spectrum beta-lactamase (ESBL) resistance in many parts of the world.1,2 The prevalence of ESBL producing E. coli (EPE) has been increasing in Denmark since the early 2000s but is now stagnating at 4–7% in blood and urine isolates.3,4 In Denmark, a study of E. coli causing UTI in general practice showed that E. coli belonging to sequence type 131 (ST131) clearly dominated the ESBL-producing population, almost always belonging to serotype O25 and, as seen in other parts of the world, most frequently carrying blaCTX-M-15.2,5,6 Hence, ESBL production seems to be correlated to specific ST and O-type in E. coli. The genes encoding ESBLs have been shown to transfer horizontally from one bacterium to another via conjugation,7 which is influenced by compatibility of the mating pair and presence of restriction/modification systems in the recipient cell.8,9 In vivo, this usually occurs in the human intestine which is a large reservoir for genetic material.10 Selective pressure from antibiotic concentrations below the minimal inhibitory concentration (MIC) has been found to affect transfer of resistance plasmids.11
In this study, we investigated the conjugational transfer of ESBL resistance encoded by blaCTX-M-15 and a possible association of conjugal frequencies to O-type and ST of the recipient. This was investigated in a wide range of fecal E. coli of different O-types and STs, including the prevalent ST131, all collected from general practices. This could reveal whether transfer of ESBL is lineage specific and merely seen in a few successful STs, like ST131, as well as predict the frequency of plasmid transfer to fecal E. coli from healthy adults. Such a study has, to our knowledge, not been performed previously with fecal E. coli of human origin as recipients.
Materials and methods
The donor isolates used in this study were E. coli collected from urine samples from general practice, previously described by Hertz et al.6 The recipient isolates were from a collection of fecal E. coli from younger women previously described by Nielsen et al.12 These isolates constituted a variety of STs, O-types and incompatibility groups (Inc-groups) (Table 1) with ampicillin MIC<8mg/L. Whole genome sequencing analyses were conducted on 29 possible donors and 19 possible recipients which were previously sequenced from Illumina paired-end and mate-pair libraries described in detail by Nielsen et al.13 Isolates sequenced in this study were sequenced (Illumina) using paired-end libraries (Nextera XT library kit) and for a subset of isolates (N=13) we were able to create mate-pair libraries (Nextera Mate Pair Sample Preparation Kit) in addition to paired-end libraries (genomic details for all isolates in Table S1). The DNA was extracted using DNeasy blood and tissue kit. Assembly was created using AllPaths-LG and the genomes were annotated with Prokka. The genomes were analyzed with Plasmidfinder 1.3, MLST 2.0 and SerotypeFinder 2.0 (all from cbs.dtu.dk) and existence of restriction/modification (R/M) systems in recipient strains was analyzed in REBASE and using GenBank followed by BLAST (80% similarity and query coverage).14 Three donor strains of different MLST type, serotypes and Inc-groups (Hvi45, Hvi132 and Hvi09) were selected for conjugation based on the genomic analyses and found to be compatible with 9, 11 and 9 recipients, respectively (Table 1). Additionally, conjugation to a laboratory K12 E. coli strain (ALO1319) was included for comparison. The recipients had no natural antibiotic drug resistance to distinguish them from the donors; thus, rifR,strR recipients were produced as described previously.15
For the conjugation, donor and recipient strains (Table 1 for performed combinations) were grown in LB broth overnight at 37°C with DNase (10 mL/L) (DNase I, Sigma Aldrich Denmark, Brøndby, Denmark) and mixed 1:1 and 100 µL was added to a sterile filter (0.22µM GSTF, Merck Millipore, Darmstadt, Germany) on a 5% blood agar plate (SSI Diagnostica, Hillerød, Denmark) and incubated overnight at 37°C. The cells were suspended from the filter in 0.9% saline by vortexing and plated in duplicates on agar plates containing rifampicin (100 mg/L)/streptomycin (100 mg/L), ampicillin (1000 mg/L) and rifampicin (100 mg/L)/streptomycin (100 mg/L)/ampicillin (1000 mg/L) (all from Sigma Aldrich, Brøndby, Denmark), respectively, after appropriate dilution in 0.9% saline. All conjugation experiments were performed in duplicates and included relevant positive and negative controls, and after 48 hrs, the plates were counted and five colonies were picked from each agar plate for verification by PCR for blaCTX-M-155 and a random amplified polymorphic DNA (RAPD) PCR16 to distinguish true transconjugants from mutated donor strains. For a subset of isolates, we performed liquid conjugation with single and double antibiotic selective plating, as described by Johnson et al.17
The plasmid transfer experiments were repeated with one donor (Hvi15) and three recipient strains (KTE72, KTE144, KTE168) under selective pressure on agar plates with ampicillin (0.75 mg/L).
Results and discussion
The aim of this study was to investigate the conjugative transfer of ESBL production caused by blaCTX-M-15 to fecal E. coli with different O-types and STs. Opposite from previous studies, this study applied recipients of fecal human origin instead of laboratory strains. All this with the purpose of indicating the likelihood of horizontal spread of ESBL resistance to other fecal E. coli, as laboratory strains have been described as poor models for in vivo plasmid transfer frequencies.18,19 The fecal isolates were prepared for conjugation by introducing not only rifampicin resistance but also streptomycin resistance to perform appropriate selection. Introduction of only one resistance marker prior to conjugation as described elsewhere17 lead to selection of rifampicin-resistant donors on the transconjugant plate (verified by RAPD PCR), whereas no donor mutants were grown on the plate when both rifampicin and streptomycin were applied (data not shown). This demonstrates the importance of double antimicrobial selection of transconjugants and molecular verification of transconjugants by, eg, RAPD PCR in order not to report falsely high conjugation frequencies.
The mate-pair genomes yielded almost complete assembly of plasmid content on the same scaffold, and from these genomes, we were able to predict whether blaCTX-M-15 was carried on a plasmid, and hence transferable, or was anchored in the chromosome unlikely to conjugate. In silico investigation identified only a few donors of ST131/O25 where blaCTX-M-15 was surrounded by plasmid material (4/12). The remaining isolates including 8 of 11 isolates belonging to the disseminating fimH30 subclone carried blaCTX-M-15 on the chromosome surrounded by transposases. Based on the genomic sequences, the three selected donors for conjugation experiments carried blaCTX-M-15 on a plasmid and not anchored in the chromosome.
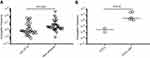
We investigated the conjugation abilities of different blaCTX-M-15-carrying donors with compatible recipients. Despite a very thorough genomic investigation for optimal donors, the results illustrate very low conjugation frequencies for all three donors (Table 1). The donors Hvi132 and Hvi09, belonging to O25/ST131 did not produce any detectable transconjugants with filter (Table 1) nor liquid conjugation (data not shown), despite carrying blaCTX-M-15 on a plasmid. The donor Hvi15 (O28/ST10) conjugated with several recipients with frequencies of 10−9–10−7/recipient and 10−10–10−7/donor (Table 1). For this donor, the median conjugation frequency was lower with recipients of O25/ST131 (KTE49, Hvi138) compared to other O- and ST-types (Figure 1A, Table 1, P=0.004). In addition, conjugation with a ST131/O25/fimH30 donor with blaCTX-M-15 situated in the genome surrounded by transposases was also investigated with several different recipients, but showed frequencies below the detection limit similar to the remaining ST131 donors (data not shown), as expected due to the genomic positioning of blaCTX-M-15. This genomic variant constituted 7 of 13 ST131/O25/fimH30 donors with high-quality mate-pair assemblies.
Conclusively, the isolates tested had lower conjugation frequencies when ST131/O25 donors or recipients were used. Several conjugation studies have been performed elsewhere using ESBL-producing E. coli strains as donors, which showed conjugation frequencies of ESBL plasmids between 10−7 and 10−2 per donor cell applying laboratory recipients.17,20,21 In these studies, only one antibiotic was applied for recipient selection and/or the post-conjugational verification of transconjugants was limited or even absent, which both could explain the different results obtained in those studies.
In the present study, the conjugative ability of blaCTX-M-15 differs with O-type and ST of the recipient. The findings indicate that E. coli O25/ST131 is a stable subtype that does not easily take up plasmids under optimal laboratory conditions. Conjugal frequencies were also low for other ST/O type donors, indicating that ESBL emergence with the presence of blaCTX-M-15 is more likely to be caused by dissemination of fit subtypes rather than by conjugal transfer of blaCTX-M-15 between E. coli. The differences in conjugation frequencies and O-type of the recipient suggest that the O-antigen could be involved in the process of conjugation. Changes in LPS have previously been found to influence conjugation frequencies, possibly by involvement in recipient cell recognition;22,23 however, further studies are necessary to enlighten this.
From a clinical aspect, the low conjugation frequencies of E. coli ST131 to clinical fecal strains are appeasing. If the widely prevalent ESBL-carrying bacterium is introduced to a host environment without antibiotic selection pressure it would, according to these results, not be likely to pass on its resistance plasmid to fecal E. coli. We cannot exclude that conjugation occurs with higher frequencies between different species in vivo. This would increase the overall gene-pool and further increase the risk of spread and infection. However, arguing against this hypothesis is our identification of blaCTX-M-15 often linked to the chromosome and not situated on a plasmid, despite being present in isolates belonging to ST131/O25/fimH30 with IncF plasmid.
Conjugation frequencies under ampicillin pressure with isolate KTE72 (O2/ST141) as recipient were 100-fold higher than frequencies from experiments without antibiotics (P=0.07) (Figure 1B). No transconjugants could be observed with recipients KTE144 (O120/ST2797) and KTE168 (O2/ST141) when subject to a sub-MIC concentration of ampicillin. This indicates that sub-lethal antibiotic pressure may not affect all E. coli conjugal frequencies, rather specific subtypes and emphasizes the importance of antibiotic stewardship programs to minimize the spread of multidrug-resistant E. coli such as O25/ST131/fimH30 with blaCTX-M-15.
The genomic characterization of the recipient isolates identified R/M-systems in all the isolates belonging to common ST types of the fecal E. coli microbiota. There was overall no correlation between number of restriction-modification systems and the conjugation frequency; however, the ST131/O25 isolates all carried at least two R/M-systems, which could perhaps explain why these isolates were poor recipients of the ESBL-carrying plasmid. We found no correlation between transfer frequencies and neither Inc-groups nor restriction/modification-systems (number and type) of the recipients. Conjugation with these clinical strains could occur even in recipients with three different R/M-systems (based on in silico analyses), supporting previous findings that presence of R/M-systems in recipients is not a complete barrier for conjugation.24 On the other hand, the presence of R/M-systems in all of the clinical fecal recipients may contribute to the generally low conjugation frequencies obtained and could be the reason why many of the conjugations were below the detection limit. Conclusively, in vivo conjugation would be challenged similarly by the abundant R/M-systems and could lower dissemination of ESBL-resistance through plasmid conjugation. The laboratory strain ALO1319 conjugated with similar frequencies despite lack of R/M-systems (Table 1).
The number of experiments done with selective pressure of ampicillin is very low. Further conjugation experiments will be necessary to uncover the effect of ampicillin on conjugation in more depth. Because of the limitations of in vitro studies, it would be very relevant to repeat the conjugation experiments in vivo, eg, in mice using clinical E. coli strains.
Conclusion
In conclusion, this is the first study to our knowledge investigating conjugal recipient abilities of clinical E. coli isolates of different sero- and ST-types. Conjugation frequencies differed with O-type and ST with significantly lower frequencies with E. coli ST131 as recipients or donor, indicating that the worldwide high prevalence of CTX-M-15-producing ST131 E. coli is not likely caused by a conjugal spread of blaCTX-M-15 to susceptible O25/ST131 strains rather due to clonal dissemination. Plasmid transfer frequencies were higher for some isolates under sub-MIC concentrations of ampicillin, which demonstrates the need for proper antibiotic stewardship programs.
Data availability
The genomic data of the recipients and donors can be found in GenBank or ENA as described in Table S1.
Ethics statement
The collection of recipient isolates was approved by the National Committee on Health Research Ethics, Denmark. The collection of the ESBL donor isolates was approved by the Danish Health Authority.
The isolates from the study by Nielsen et al.12 were collected with informed consent. For the donors previously described by Hertz et al.6 the Danish health Authority approved the E. coli collection without collecting informed consent. These isolates were collected from routine diagnostics on UTI samples sent from the patient’s own general practitioner to the regional hospital for diagnostics. The collection was performed at the regional hospital.
Acknowledgments
This work was supported by Danish Council for Independent Research and by MICA Foundation. Funding sources had no influence on study design, data interpretation or the final paper.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gunther NW, Snyder JA, Lockatell V, Blomfield I, Johnson DE, Mobley HLT. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect Immun. 2002;70(7):3344–3354. doi:10.1128/iai.70.7.3344-3354.2002
2. Livermore DM, Canton R, Gniadkowski M, et al. CTX-M : changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59:165–174. doi:10.1093/jac/dkl483
3. DANMAP. DANMAP 2015 - Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark. Denmark: 2015.
4. Brolund A. Overview of ESBL-producing enterobacteriaceae from a nordic perspective. Infect Ecol Epidemiol. 2014;4:1–9.
5. Olesen B, Hansen DS, Nilsson F, et al. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J Clin Microbiol. 2013;51(6):1779–1785. doi:10.1128/JCM.00346-13
6. Hertz FB, Nielsen JB, Schønning K, et al. Population structure of drug-susceptible, -resistant and ESBL-producing Escherichia Coli from community- acquired urinary tract. BMC Microbiol. 2016;16(63):1–6. doi:10.1186/s12866-015-0617-z
7. Mshana SE, Imirzalioglu C, Hossain H, Hain T, Domann E, Chakraborty T. Conjugative IncFI plasmids carrying CTX-M-15 among Escherichia coli ESBL producing isolates at a university hospital in Germany. BMC Inf Dis. 2009;9(97):8. doi:10.1186/1471-2334-9-97
8. Mruk I, Kobayashi I. To be or not to be : regulation of restriction – modification systems and other toxin – antitoxin systems. Nucleic Acids Res. 2014;42(1):70–86. doi:10.1093/nar/gkt711
9. Novick RP. Plasmid Incompatibility N. Microbiol Rev. 1987;51(4):381–395.
10. Huddleston JR. Horizontal gene transfer in the human gastrointestinal tract : potential spread of antibiotic resistance genes. Infect Drug Resist. 2014;7:167–176. doi:10.2147/IDR.S48820
11. Schuurmans J, van Hijum SA, Piet JR, et al. Effect of growth rate and selection pressure on rates of transfer of an antibiotic resistance plasmid between E. coli strains. Plasmid. 2014;72:1–8. doi:10.1016/j.plasmid.2014.01.002
12. Nielsen KL, Dynesen P, Larsen P, Frimodt-Møller N. Faecal Escherichia coli from patients with E. coli urinary tract infection and healthy controls who have never had a urinary tract infection. J Med Microbiol. 2014;63(Pt 4):582–589. doi:10.1099/jmm.0.068783-0
13. Nielsen KL, Stegger M, Kiil K, et al. Whole-genome comparison of urinary pathogenic Escherichia coli and faecal isolates of UTI patients and healthy controls. Int J Med Microbiol. 2017;307(8):497–507. doi:10.1016/j.ijmm.2017.09.007
14. Roberts RJ, Vincze T, Posfai J, Macelis D, Biolabs NE. REBASE –– a database for DNA restriction and modification : enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi:10.1093/nar/gku1046
15. Baquero M, Galan JC, Turrientes M, et al. Increased mutation frequencies in Escherichia coli isolates harboring extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2005;49(11):4754–4756. doi:10.1128/AAC.49.2.612-618.2005
16. Nielsen KL, Godfrey PA, Stegger M, Andersen PS, Feldgarden M, Frimodt-Møller N. Selection of unique Escherichia coli clones by random amplified polymorphic DNA (RAPD): evaluation by whole genome sequencing. J Microbiol Methods. 2014;103:101–103. doi:10.1016/j.mimet.2014.05.018
17. Johnson TJ, Danzeisen JL, Youmans B, et al. Separate F-type plasmids have shaped the evolution of the H 30 subclone of escherichia coli sequence type 131. mSphere. 2016;1(4):e00121–16. doi:10.1128/mSphere.00121-16
18. Hobman JL, Penn CW, Pallen MJ. Laboratory strains of Escherichia coli : model citizens or deceitful delinquents growing old disgracefully? MicroOpinion. 2007;64:881–885.
19. Gordon D. Rate of plasmid transfer among Escherichia coli strains isolated from natural populations. J Gen Microbiol. 1992;138:17–21. doi:10.1099/00221287-138-1-17
20. Warnes SL, Highmore CJ, Keevil CW, Bassler B. Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces : implications for public health. Mbio. 2012;3(6):1–10. doi:10.1128/mBio.00489-12
21. Vk V. Horizontal transfer of antimicrobial resistance by extended-spectrum β lactamase-producing enterobacteriaceae. J Lab Physicians. 2011;3(1):37–42. doi:10.4103/0974-2727.78563
22. Anthony KG, Sherburne C, Sherburne R, Frost LS. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994;13(6):939–953.
23. Sherburne C, Taylor DE. Effect of lipopolysaccharide mutations on recipient ability of salmonella typhimurium for incompatibility group H plasmids. J Bacteriol. 1997;179(3):952–955. doi:10.1128/jb.179.3.952-955.1997
24. Roer L, Aarestrup FM, Hasman H. The EcoKI type i restriction-modification system in Escherichia coli affects but is not an absolute barrier for conjugation. J Bacteriol. 2015;197(2):337–342. doi:10.1128/JB.02418-14
Supplementary material
 |
Table S1 Genomic data on donors and recipients including sequencing method, mean coverage, number of contigs/scaffolds, genome size, accession number |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.