Back to Journals » Infection and Drug Resistance » Volume 12
Epidemiology of Plasmodium falciparum infection and drug resistance markers in Ota Area, Southwestern Nigeria
Authors Olasehinde GI , Diji-Geske RI , Fadina I, Arogundade D , Darby P, Adeleke A, Dokunmu TM , Adebayo AH, Oyelade J
Received 10 October 2018
Accepted for publication 13 March 2019
Published 5 July 2019 Volume 2019:12 Pages 1941—1949
DOI https://doi.org/10.2147/IDR.S190386
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
GI Olasehinde,1 RI Diji-Geske,1 I Fadina,1 D Arogundade,1 P Darby,1 A Adeleke,1 TM Dokunmu,2 AH Adebayo,2 J Oyelade3
1Department of Biological Sciences; 2Department of Biochemistry; 3Department of Computer and Information Sciences, Covenant University, Ota, Ogun State, Nigeria
Purpose: Effective routine monitoring and surveillance of parasite genes is a necessary strategy in the control of parasites’ resistance to antimalarial drugs, according to the WHO’s recommendation. This cross-sectional study therefore aimed at carrying out an epidemiological analysis on malaria incidence in Ado-Odo/Ota, Ogun State.
Patients and methods: Blood and corresponding saliva samples were collected from 1,243 subjects of all ages and sex presenting with fever and a parasitemia level ≥2,000 between September 2016 and March 2018. Samples were collected from selected health facilities in the study area of Ogun state to establish the prevalence of falciparum malaria and determine resistance genes harbored by the parasites. The overall prevalence of falciparum malaria in the study site by microscopic examination was 45.86%. The highest incidence of 57.42% was recorded among male subjects. Point mutations of K76T and N86Y in the Pfcrt and pfmdr-1 genes, as well as non-synonymous mutations in Pfk13 genes, were screened for and sequenced for further analysis.
Results: Pfcrt was detectable in 57.42% of blood and 51.02% of saliva samples, respectively. About 34.78% of the subjects that were confirmed microscopically harbored the Pfmdr-1 mutated gene while 26.67% of the saliva samples revealed Pfmdr-1. Epidemiological studies identified the presence of wild-type Pfk13 genes in 21.84% of blood and 44.44% of saliva samples correspondingly. For each of the genes evaluated, saliva portrayed great diagnostic performance when compared with blood.
Conclusion: Findings from this study have established the prevalence of malaria and the resistance pattern of P. falciparum in the study area. The findings may help in formulating drug policies and suggest the use of saliva as a noninvasive point-of-care method of diagnosing malaria potentially deployable to rural endemic areas.
Keywords: prevalence, resistance, malaria, Ado-Odo/Ota
Introduction
Malaria, the life-threatening vector-borne disease, remains a global threat and a major public health issue across the world.1 About 219 million cases and 435,000 deaths from malaria occurred worldwide in 2017.2 A major factor hindering malaria elimination strategies is the continuous emergence of Plasmodium falciparum parasites resistant to all known existing malaria drugs.3,4 However, according to the WHO, malaria can be prevented and even cured, and amplified efforts between the years 2000 and 2015 have considerably decreased the fatality rate of malaria by 60%.1
In retrospect to all intensified control methods against malaria, drug resistance has remained on the rise, hence posing a serious challenge where control and prevention are concerned and increasing the need for vaccine development.4 Antimalarial drug resistance may occur due to variations in the build-up of drug or efflux mechanisms (as in the case of chloroquine, amodiaquine, quinine, halofantrine, and mefloquine resistance) or due to diminished affinity of the drug’s target, which may result from point mutations in the respective genes that encode these targets (pyrimethamine, cycloguanil, sulphonamide, atovaquone, and artemisinin resistance).5 Following the resistance of Plasmodium falciparum to antimalarial drugs such as chloroquine and sulfadoxine-pyrimethamine, the WHO in 2001 endorsed artemisinin combination therapies (ACTs) comprising drug combinations of artemether-lumefantrine (AL) and amodiaquine-artesunate (AQ-AS) as first-line treatment for uncomplicated malaria.4 The efficacy of artemisinin combination drugs has been proven,6 notwithstanding drug combinations have a lower risk of resistance and may present greater problems if such occurs.
All existing malaria diagnostic measures (eg, PCR, microscopy, blood smear, and RDTs) involve the use of blood through finger pricking or drawing with needle and syringe. These invasive methods of malaria diagnosis are not without discouraging factors such as pain associated with needles, fear of contraction of an infectious disease like HIV, high cost of diagnosis, traditional bias and so on, hence leading to self-medication and eventually resistance. Based on these, other sources of body fluid such as human saliva are being explored to determine their potential as a malaria diagnostic tool. This is crucial in the battle against resistance in order to enhance the speedy detection of parasites upon symptoms. Human saliva is already being used to detect a broad band of diseases such as cancers and cardiovascular diseases due to its constituents of proteins, electrolytes, and DNA.7 Viral infections such as HIV and human papillomavirus (HPV) have also been detected using saliva diagnostics test kits.8,9 Some studies have reported the detection of Plasmodium falciparum in the saliva of infected subjects.10,11 Therefore, saliva may be a more encouraging noninvasive substitute for malaria diagnosis other than blood.12
Antimalarial drug resistance can be evaluated in vivo and in vitro by parasite susceptibility or by the exploitation of molecular methods such as PCR techniques to identify genetic markers. Drug resistance can lead to the inability to clear asexual parasites found in the peripheral blood, which eventually develop into gametocytes, ie, the transmission stage of the parasite genotype.5 PCR methods are principally useful for findings on strain variation, mutations, and examination of parasite genes involved in drug resistance. In the battle against malaria, the identification of the gene responsible for chloroquine resistance transporter (Pfcrt) brought about a scientific revolution.13 Between the amino acids lysine and threonine lie other polymorphic amino acids; however, a change in the Pfcrt codon K76T gene presents as the most dependable genetic marker among them all and may be essential for coding the resistance makeup.14–16 Notwithstanding, chloroquine resistance could occur in mutations anywhere between codons 72 and 76. P. falciparum multidrug resistance (Pfmdr-1) occurs due to a substitution in codon N86Y chromosome and is associated with drug resistance to chloroquine, halofantrine, quinine, and mefloquine. Some studies identified the association of single nucleotide polymorphism (SNP) causing a base change from asparagine to tyrosine at codon 86 (N86Y) with chloroquine resistance both in vitro and in vivo.17–19
Existing malaria diagnostic methods using either rapid diagnostic test (RDT) and/or microscopy require the use of blood samples for effective diagnosis. Although these approaches are sensitive, accurate, relatively cheaper and rapid, their greatest disadvantage is their dependency on blood samples for diagnosis due to the increased risks of needle injuries and risk of accidental infection from diseases which are common in malaria-endemic areas as well as cultural blood taboos in certain African communities. These serve as limitations, particularly in longitudinal studies that require repeated testing and monitoring to be conducted, as it reduces community participation.20
The use of body fluids such as saliva has been employed as a diagnostic test fluid for determining immunity and detecting diseases such as cancers, cardiovascular diseases, HIV, etc.21–23 It is convenient to use for diagnosis as it is not difficult to collect, noninvasive, and possesses minimal risk of infection compared to blood collection. Saliva contains a minimum of 400 proteins which interface with the salivary glands and blood vessels, exchanging materials at intervals.24,25 Diffusion and active transport of human pathogens are therefore most likely to end up in the saliva, hence presenting a reliable tool for disease diagnosis.7 Moreover, the reliability of PCR as a malaria detection tool using saliva could be optimized and remains consistently applicable across various populations.
Artemisinin-based combination therapies recommended by WHO are effective against Plasmodium parasites and can curb the spread of resistance.26 However, the risk of developing resistance to combined drugs is now evident from Southeast Asia, where ACT resistance has now emerged.27 As the parasite develops multi-drug resistance to a previously active drug, it threatens malaria control in endemic areas which largely depend on chemotherapy. The quest to determine the presence of parasite resistance to ACTs and other antimalarial drugs including artemisinin and its derivatives which is the hallmark for treatment of malaria, is the drive for carrying out this study and aimed at determining the development of new methods for malaria diagnosis. Notwithstanding important gains in fields of research in malaria, its methods of diagnosis remain invasive, with the drugs for treatment not available in many rural communities, and if available may be taken in inappropriate doses, thus leading to further resistance. Furthermore, effective monitoring of parasite-resistant genes is necessary for determining the efficacy of currently used regimens and deciding on the time point for introducing new drugs/re-introducing old drugs. It is also crucial in the development of new diagnostic tools, such as saliva body fluid, which will aid in curbing malaria transmission as well as preventing resistance. Hence, this study is focused on carrying out an epidemiological study on falciparum malaria infection in Ado-Odo/Ota, Ogun State, Nigeria and determine the prospect using of saliva in detecting P. falciparum markers of resistance.
Materials and methods
Infected human blood and corresponding saliva samples were obtained from selected hospitals in the study site. Males and females of different age ranges presenting with fever and body temperature greater than 37°C were recruited to the study. The study was conducted in accordance with the Declaration of Helsinki, and ethical permission for this study was approved by the Covenant University Biological Sciences Ethical Review Committee (CUBIOSCREC) and Covenant Health Research Ethics Committee (CHREC). All research participants approved their informed consent and minors were represented by their guardians. Inclusion criteria included persons willing to complete and sign the consent form and with significant parasitemia level of ≥2,000 parasites/µL of blood.
Thin and thick films of the blood samples were stained with Giemsa stain. Blood films were examined microscopically using the 100× (oil immersion) objective lens. Qiagen DNA extraction kit (Qiagen NV, Venlo, the Netherlands) was used to extract Plasmodia DNA from the blood and saliva samples. NanoDrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to check for DNA concentration and purity using a ratio of 260/280. Point mutations in the genes of Pfcrt and Pfmdr-1 were analyzed using PCR/nested PCR-restriction fragment length polymorphism (RFLP) methods,14 and used to amplify and confirm the mutation of resistant genes. If any discrepancies were found by different diagnostic methods, those samples were excluded from the study. Afterward, the amplicons were run electrophoretically on a gel to check for bands size of the amplified resistance gene region.
The data obtained were analyzed using Microsoft Excel. Statistical analysis was carried out using P-values from a chi-square test for proportions using a P-value of 0.05, comparing the relationship between age and rate of incidence of falciparum malaria. Values less than 0.05 were accepted as statistically significant. The analysis was done using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
The results of the epidemiological study of Plasmodium falciparum infection in Ado-Odo/Ota Local Government Area of Ogun State, Southwestern Nigeria in 1,243 subjects are presented in Tables 1–5. The malaria incidence detected by microscopy in 2017 was 52.09% with the highest incidence of 55.38% among the age group ≤5 years (Table 1). Table 2 shows the incidence of malaria among study subjects in 2018, with an overall incidence of 44.11%. Incidence was higher among males within the same age range in this study. The overall prevalence of falciparum malaria during the study period was 45.86% (Table 3). There was a significant association between age younger than 20 years and the incidence of malaria during the study as shown in Table 4. The prevalence of the different molecular markers screened from blood and corresponding saliva samples in the subjects are presented in Table 5. Figure 1 shows the overall detection of malaria in saliva and blood in all samples; these rates were similar. Plates 1–5 show the band sizes of the amplified molecular markers of antimalarial resistance on an agarose gel.
 |
Table 1 Incidence of falciparum malaria in the study area (2017) |
 |
Table 2 Incidence of falciparum malaria in the study area (2018) |
 |
Table 3 Prevalence of falciparum malaria in the study area (2017/2018) |
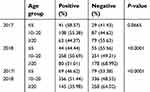 |
Table 4 Chi-square analysis showing the relationship between the incidence of falciparum malaria and age |
 |
Table 5 Prevalence of resistance genes in Ado-Odo/Ota, Ogun state |
 |
Figure 1 Detection of P. falciparum resistance genes in blood and saliva. |
 |
Plate 1 Gel electrophoresis result of the nested PCR product carried out on blood and saliva showing the Pfcrt genetic marker. Abbreviations: M, molecular marker; bp, base pair. |
 |
Plate 2 Gel electrophoresis result of the nested PCR of blood samples showing the Pfcrt genes. |
 |
Plate 3 Gel electrophoresis result of the nested PCR on blood samples for Pfmdr1 genetic marker. |
 |
Plate 4 Gel electrophoresis result of the nested PCR product carried out on blood samples showing the K13 genetic marker. |
 |
Plate 5 Gel electrophoresis result of the nested PCR product carried out on saliva samples 1–11 showing the K13 genetic marker. Positive bands: Lane 2-3-7-8-9-10-11. |
Discussion
A key instrument to successful management of the global malaria challenge remains the WHO’s recommended continuous monitoring and effective surveillance of resistance development in the parasites’ genes.27 This study presents the epidemiology of falciparum malaria in Ado-Odo/Ota, Ogun State, Nigeria, and surveillance for resistance markers using blood and saliva, a noninvasive sampling method. In the relatively large samples evaluated in the study area, malaria incidence was over 45% and increased over the two-year period, which suggests that malaria transmission is sustained in an era of malaria elimination. Global reports on malaria control indicate stalled progress in the successful reduction of malaria transmission, especially for Nigeria, which has the highest burden of malaria cases globally and in Africa, followed by Madagascar and the Democratic Republic of the Congo, where increases in malaria rates from 2016 to 2017 were recorded.28 Our findings lend support to this and further suggest that this increasing trend appears to continue into 2018.
The incidence of falciparum malaria following microscopy in 2017 was 52.09% with the highest rate of 55.38% among the age range 10–20 years (Table 1). Table 2 shows the incidence of malaria among study subjects in 2018, with an incidence of 44.11%. It was observed that incidence was higher among males within this age range in this study. The reason for this may be factors such as habits and lifestyles associated with males within that age bracket, some of which may include negligence in the intermittent use of bed nets, greater exposure to mosquito bites from play areas and drugs misuse. Females on the other hand exhibit carefulness in these areas. Age was also shown to be a risk factor for malaria,1 as the incidence of falciparum malaria considerably increased with age. The overall prevalence of falciparum malaria during the study period was 45.86% (Table 3). The incidence in 2018 and overall prevalence show that there is a significant association between age and incidence of malaria in the study as shown in Table 4.
Artemisinin and ACT resistance has developed in the Greater Mekong area of Southeast Asia (GMS) and has continued to spread with increasing drug failure to artemisinin combination therapies (ACTs); however, across Africa, diverse non-synonymous K13 mutations are being reported,27 and non-artemisinin resistance markers have been reported to be associated with ACT response,14–19 and of recent to partial artemisinin resistance but in the African region no study has attributed this to K13 mutations.27 The strategy to contain this spread thus warrants continued surveillance for these markers. Point mutations in codons K76T and N86Y of Pfcrt and Pfmdr-1 genes, respectively, as well as non-synonymous mutations in Pfk13 genes, were targeted using PCR/nested PCR-RFLP and sequenced for further analysis.
We identified the presence of Plasmodium falciparum chloroquine resistance transporter gene mutations in 57.42% of 58 blood and 51.02% of 25 saliva samples. Plasmodium falciparum multidrug resistance gene mutations were detected in 34.78% of 16 blood samples and 26.67% of eight saliva samples evaluated while Pfk13 gene was amplified in 21.84% of 19 blood samples and 44.44% of eight saliva samples (Figure 1). Sequencing analysis revealed no mutations in the K13 propeller domain, but various studies have shown that the African K13-propeller mutations vary from those found in Southeast Asia, and partial artemisinin resistance in sub-Saharan Africa is developing.27–31 Several independent mutations developed in the GMS in the PfK13 propeller domain in the Southeast Asian parasites; compared to those of African origin, these genetic changes are less likely to be widespread due to high transmission rates because they will be lost to outcrossing; however, historically, resistance spreads from Asia to Africa, where the higher propagation limits elimination.
Mutations in Pfcrt K76T codon have commonly been associated with chloroquine resistance, while mutations in Pfmdr-1 N86Y have been associated with both chloroquine and amodiaquine resistance.14–19,31 High rates of Pfcrt K76T were detected both in blood (47.89%) and saliva (31.43%). This study presented results that attempt to identify the presence of these gene polymorphisms in saliva of humans with amplifiable malarial DNA using nested PCR. This indicates that there is a possibility that Plasmodium falciparum DNA is introduced into the human saliva via an intracellular component; probably through red blood cells that have been ruptured or DNA from parasites trapped in macrophages.32 Hence, challenges limiting the implementation of interventions requiring country-wide screening for periodic malaria surveillance, which is more effective for eradication of malaria in other regions, can now be overcome using saliva as a diagnostic fluid.
The incidence of Plasmodium falciparum malaria from blood and saliva samples was established with the amplification of parasite genomic DNA from blood and saliva samples, and point mutations in Pfcrt, Pfmdr-1 and P. falciparum kelch propeller gene molecular marker were also detected. Despite the fact that human saliva is mostly composed of water (98%), it contains lots of electrolytes, proteins, and DNA, and it has been and is currently being explored for the monitoring and diagnosis of a wide variety of human diseases.12 This study reveals the potential of saliva as a reliable diagnostics tool for malaria detection and for mapping the spread of resistance in this era of malaria eradication, hence making it easier to overcome the reversal of decreasing transmission intensities in high malaria endemic regions such as Nigeria.
Prevailing factors such as fear of needles, risk of infections and perhaps traditional taboos may serve as a deterrent to the successful diagnosis of malaria, especially in children and among rural dwellers. However, most commonly used malaria diagnoses for epidemiological surveys rely on the use of blood drawn using needles, which inflicts pain on the subject. Because young children are the most vulnerable to infection and disease, they tend to constitute the de facto sentinel group used for most malaria surveys. Their unwillingness to participate in epidemiological survey owing to the invasiveness of the diagnosis may grossly mitigate malaria surveillance and control. This study showed high detection rates comparable to blood diagnosis of molecular markers of antimalarial resistance in the study area and reported the absence C580Y or A578S SNP; the most frequent K13 allele observed in Africa, associated with artemisinin resistance in sub-Saharan African parasites and those found in Southeast Asia.30,31
Conclusion
This research further analyzed the incidence and prevalence of Plasmodium falciparum in Ado-Odo/Ota Local Government area of Ogun State.2 The data provided reveal high endemicity of falciparum malaria, the existence of malaria resistance genes in the study area and the prospect of saliva as a noninvasive “point-of-care” diagnostics tool. It also confirms that subjects with lower age range are more predisposed to falciparum malaria. The research achieved and encourages systematic monitoring and surveillance of malaria parasite infection caused by P. falciparum in the study area which is in line with the WHO’s recommendation for effective routine monitoring.1 This will aid in providing an updated guide in the use of ACTs.
Disclosure
The authors report no conflicts of interest in this work.
References
1. World Health Organization. World Malaria Report. Geneva: WHO; 2015. Available from: http://www.who.int/malaria/publications/world-malaria-report-2015/report/en/.
2. Diji-geske R, Olasehinde G, Fadina I, Arogundade D, Darby P. Epidemiological data of falciparum malaria in Ado-Odo/Ota, Southwest Ogun State, Nigeria. Data Brief. 2018;19:1398–1402.
3. Olasehinde GI. In- Vitro and Molecular Studies on the Resistance of P. Falciparum to Antimalarial Drugs in Ogun State, south- western Nigeria. Lambert Academic publishing; 2010. 978-3-8433-7825-3.Available from: http://eprints.covenantuniversity.edu.ng/24/2/Olasehinde_phdwork_%281%29.pdf.
4. Olasehinde GI, Ojurongbe DO, Akinjogunla OJ, Egwari LO, Adeyeba AO. Prevalence of malaria and predisposing factors to antimalarial drug resistance in Southwestern Nigeria. Res J Parasitol. 2015;10(3):92–101. doi:10.3923/jp.2015.92.101
5. Shweta S, Bikash M, Rakesh S. Challenges of drug resistant malaria. Parasite. 2014;21:61. doi:10.1051/parasite/2014059
6. Olliaro PL, Taylor WRJ. Antimalarial compounds: from bench to bedside. J Exp Biol. 2003;206:3753–3759.
7. Pfaffe T, Cooper-White J, Beyerlein P, Kostner K, Punyadeera C. Diagnostic potential of saliva: current state and future applications. Clin Chem. 2011;57(5):675–687. doi:10.1373/clinchem.2010.153767
8. Zachary D, Mwenge L, Muyoyeta M, et al. Field comparison of OraQuick ADVANCE rapid HIV-1/2 antibody test and two blood-based rapid HIV antibody tests in Zambia. BMC Infect Dis. 2012;12:183. doi:10.1186/1471-2334-12-183
9. SahebJamee M, Boorghani M, Ghaffari SR, Atarbashi MF, Keyhani A. Human papillomavirus in saliva of patients with oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2009;14:525–528. doi:10.4317/medoral.14.e525
10. Ghayour NZ, Oormazdi H, Akhlaghi L, et al. Detection of Plasmodium vivax and Plasmodium falciparum DNA in human saliva and urine: loop-mediated isothermal amplification for malaria diagnosis. Acta Trop. 2014;136:44–49. doi:10.1016/j.actatropica.2014.03.029
11. Putaporntip C, Buppan P, Jongwutiwes S. Improved performance with saliva and urine as alternative DNA sources for malaria diagnosis by mitochondrial DNA-based PCR assays. Clin Microbiol Infect. 2011;17:1484–91.32. doi:10.1111/j.1469-0691.2011.03507.x
12. Kenji OM, Samuel TY, Livo FE, et al. Detection of plasmodium falciparum DNA in saliva samples stored at room temperature: potential for a non-invasive saliva-based diagnostic test for malaria. Malar J. 2017;16:434. doi:10.1186/s12936-017-2084-5
13. Fidock DA, Nomura T, Cooper RA, Su X, Talley AK, Wellems TE. Allelic modifications of the cg2 and cg1 genes do not alter the chloroquine response of drug-resistant Plasmodium falciparum. Mol Biochem Parasitol. 2000;110:1–10. doi:10.1016/S0166-6851(00)00249-8
14. Djimde A, Doumbo OK, Cortese JF, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi:10.1056/NEJM200101253440403
15. Plowe CV, Roper C, Barnwell JW, et al. World Antimalarial Resistance Network (WARN) III: molecular markers for drug resistant malaria. Malar J. 2007;6:121. doi:10.1186/1475-2875-6-121
16. Ojurongbe O, Ogungbamigbe TO, Fagbenro-Beyioku AF, Fendel R, Kremsner PG, Kun JF. Rapid detection of Pfcrt and Pfmdr1 mutations in Plasmodium falciparum isolates by FRET and in vivo response to chloroquine among children from Osogbo, Nigeria. Malar J. 2007;6:41. doi:10.1186/1475-2875-6-41
17. Pickard AL, Wongsrichanalai C, Purfield A, et al. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–2423. doi:10.1128/AAC.47.8.2418-2423.2003
18. Basco LK, Ringwald P. Analysis of the key Pfcrt point mutation and in vitro and in vivo response to chloroquine in Yaounde, Cameroon. J Infect Dis. 2001;183:1828–1831. doi:10.1086/320726
19. Dorsey G, Kamya MR, Singh A, Rosenthal PJ. Polymorphisms in the Plasmodium falciparum Pfcrt and Pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J Infect Dis. 2001;183:1417–1420. doi:10.1086/319865
20. Wilson NO, Adjei AA, Anderson W, Baidoo S, Stiles JK. Detection of Plasmodium falciparum histidine-rich protein II in saliva of malaria patients. Am J Trop Med Hyg. 2008;78(5):733–735. doi:10.4269/ajtmh.2008.78.733
21. Oba IT, Spina AM, Saraceni CP, et al. Detection of hepatitis A antibodies by ELISA using saliva as clinical samples. J Inst Trop Med São Paulo. 2000;42:197–200. doi:10.1590/S0036-46652000000400004
22. Thieme T, Piacentini S, Davidson S, Steingart K. Determination of measles, mumps and rubella immunization status using oral fluid samples. J Am Med Assoc. 1994;272:219–221. doi:10.1001/jama.1994.03520030061029
23. Hunt AJ, Connell J, Christofinis G, et al. The testing of saliva samples for HIV-1 antibodies: reliability in a non-clinic setting. Genitourin Med. 1993;69:29–30. doi:10.1136/sti.69.1.29
24. Lima DP, Diniz DG, Moimaz SA, Sumida DH, Okamoto AC. Saliva: reflection of the body. Int J Infect Dis. 2010;14(3):184–188. doi:10.1016/j.ijid.2009.04.022
25. Pooe OJ, Shonhai A, Mharakurwa S. A PCR screen for malaria carrier infections using human saliva samples. Afr J Microbiol Res. 2011;5(28):5120–5126.
26. Vicki S. Malaria, A Global Challenge. Society for General Microbiology. Available from: https://microbiologyonline.org/file/7416093e224285db89c8ae9761d9f53f.pdf.
27. World Health Organisation. Status Reports on Artemisinin Resistance and ACT Efficacy. WHO/CDS/GMP/2018.18.
28. World Health Organisation(2018). World Malaria Report 2018. Geneva. CC BY-NC SA 3.0 IGO. ISBN 978-92-4-156565-3.
29. Lucie P, Ramadani A, Mercereau-Puijalon O, Augereau J, Benoit-Vical F. Plasmodium falciparum: multifaceted resistance to artemisinins. Malar J. 2016;15:149. doi:10.1186/s12936-016-1206-9
30. Conrad MD, Bigira V, Kapisi J, et al. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. Public Lib Sci. 2014;9(8):e105690.
31. Kaufman E, Lamster IB. The diagnostic application of saliva - a review. Oral Biol Med. 2002;13:197–212.
32. Taylor SM, Parobek CM, DeConti DK, et al. Absence of putative plasmodium falciparum artemisinin resistance mutations in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;211:1680–1688. doi:10.1093/infdis/jiu467
33. Edwin PM, Chinweizu EU, Mark IM. Amodiaquine and Ciprofloxaxin combination in plasmodiasis therapy. J Trop Med. 2015;15(1):1–10.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
