Back to Journals » Cancer Management and Research » Volume 12
ENST00000430471 Promotes Development and Metastasis of Colorectal Cancer by Regulating the Expression of YBX-1
Authors Zhu Z, Zhang X, Zhou Y, Cheng J, Xu Z
Received 25 May 2020
Accepted for publication 23 July 2020
Published 11 August 2020 Volume 2020:12 Pages 7189—7197
DOI https://doi.org/10.2147/CMAR.S264308
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xueqiong Zhu
Zhenghai Zhu,* Xiaoxin Zhang,* Ying Zhou, Jie Cheng, Zipeng Xu
Department of General Surgery, Xishan People’s Hospital, Wuxi, Jiangsu 214000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zipeng Xu Email [email protected]
Background Information: Colorectal cancer (CRC) is a common malignant tumor of the digestive system. Long non-coding (lnc) RNA ENST00000430471 has been reported to be involved in CRC development and metastasis because of its cancer-promoting ability. However, the detailed molecular mechanisms of ENST00000430471 in CRC remain largely unknown.
Methods: The cell proliferation assay and Xenograft experiment were performed to examine the proliferation rate of the tumor cells. Invasiveness and migration capability of the cells were evaluated using the invasion and wound healing assays, respectively. In addition, the RNA pull-down assay and subsequent mass spectrometry techniques were performed to identify the proteins interacting with ENST00000430471.
Results: Herein, small specific inhibiting (si) RNA-si0471#2 was used to silence ENST00000430471 in HCT116 and SW620 cell lines. This led to a significant reduction in cell proliferation, migration, and invasion. The RNA pull-down assay and mass spectrometry further revealed that ENST00000430471 interacted with several proteins such as the Y-box-binding protein 1 (YBX-1). On one hand, silencing of ENST00000430471 decreased the mRNA and protein expression levels of YBX-1. On the other hand, overexpression of YBX1 partially attenuated the suppression of cell proliferation, invasion, and migration induced by ENST00000430471 silencing.
Conclusion: Silencing of ENST00000430471 inhibits proliferation, migration, and invasion of CRC cells by regulating YBX-1 expression. These results provide baseline information that is essential in the identification of effective therapeutic targets for CRC therapy.
Keywords: lncRNA, ENST00000430471, progress, colorectal cancer, YBX-1
Introduction
Colorectal cancer (CRC) is a common malignant neoplasm characterized by high morbidity and mortality rates.1 Although there has been progress in CRC treatment, more than 190,000 patients succumb to CRC every year in China because of the large population base.2 Metastasis is one of the primary causes of CRC treatment failures that leads to death.3 Lymphatic metastasis is the most common cause of CRC metastasis. It can significantly reduce the 5-year survival rate of CRC patients.4 Cognizant to this, it is important to explore the underlying molecular mechanisms involved in the development of CRC.
Long non-coding (lnc) RNAs (length, >200 nucleotides) are a group of functional RNA molecules with limited protein-coding potential.5 They can regulate multiple biological processes such as proliferation, differentiation, apoptosis, invasion and metastasis.6 Previous studies have revealed that several lncRNAs such as CCAT1, CCAT2, HOTAIR and H19 play significant roles in the development of CRC. However, the mechanism of action of lncRNAs in lymphatic metastasis is not yet clear.7–10
In a recent study, ENST00000430471 was found to be significantly upregulated in the metastatic lymph node tissues than in paired tumor tissues and normal tissues. Moreover, overexpression of ENST00000430471 promoted CRC cell proliferation and metastasis.11 Herein, silencing of ENST00000430471 inhibited cell proliferation, migration, and invasion in HCT116 and SW620 cells. RNA pull-down assay further revealed that ENST00000430471 interacted with the Y-box-binding protein 1 (YBX-1). Overexpression of YBX-1 partially restored the tumor inhibitory effects induced by ENST00000430471 silencing.
Materials and Methods
Cell Culture
Human HCT116 and SW620 CRC cell lines were sourced from the Cell Bank of the Chinese Academy of Medical Sciences. They were then cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin (Thermo Fisher Scientific, Inc.) and incubated in a humidified incubator with 5% CO2 at 37°C.
RNA Extraction and Reverse Transcription-Quantitative PCR (RT-qPCR) Analysis
Total RNA was extracted from cultured cells using the TRIzol® reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. It was then reverse transcribed into cDNA using the PrimeScriptTMRT reagent kit (Takara Biotechnology Co., Ltd.) and used for qPCR analysis. qPCR was performed using SYBR Green fluorescent DNA binding dye (Takara Biotechnology Co., Ltd.) according to the manufacturer’s instructions. qPCR results were normalized to GAPDH. Primers used are displayed in Table 1.
 |
Table 1 Primers Used in Reverse Transcription-Quantitative PCR (RT-qPCR) Analysis |
Western Blot Analysis
Protein lysates were extracted from the cells during their logarithmic growth phase using the radioimmunoprecipitation assay (RIPA) lysis buffer. They were then separated using 10% Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the resultant bands transferred onto polyvinylidene difluoride membranes. The membranes were then blocked with 5% bovine serum albumin for 1 hour at 37°C and then incubated with primary antibodies at 4°C for 12 hours. The membranes were then washed with TBST to remove the excess antibodies and subsequently incubated with the secondary antibody, and the proteins were visualized using enhanced chemiluminescence. The following primary antibodies were used: anti-YBX1 (Abcam), and anti-β-ACTIN (Abcam).
Cell Transfection
Small interfering (si) RNAs against ENST00000430471 (si0471#1, 2 and 3), negative control (NC) siRNA (siNC), short hairpin (sh) RNA against ENST00000430471 (sh0471) and control shRNA (shNC) were purchased from Shanghai GenePharma Co. Limited. Similarly, YBX-1 expression vector (pcDNA-YBX-1) was constructed by Shanghai GenePharma Co. Limited. Transfection of the aforementioned siRNA and pcDNA was done using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. Two mL serum-free medium and an appropriate amount of lentivirus containing either sh0471 or shNC were added to each well of a 6-well plate containing the cells. The medium was replaced with fresh medium after 4–6 hours. The cells were transfected for 24–48 hours and then collected for determination.
Cell Proliferation Assay
The proliferation rate of HCT116 and SW620 cells was assessed using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay reagent in accordance with the manufacturer’s instructions. Cell proliferation rate was measured at 12, 24, 48, 60 and 72 hours after transfection. The absorbance of cells in each well was measured at a wavelength of 450 nm using a microplate reader. The assay was done in triplicate.
Xenograft Experiment
Four weeks old BALB/c nude mice were used for these experiments. They were purchased from the Model Animal Research Center and maintained under special pathogen-free conditions prior to the experiment. Their beddings and food were changed every 2 days. Moreover, their health condition was evaluated by keeping track of their appetite, reaction and breathing. Two groups of mice each comprised of 10 randomly selected mice were subcutaneously injected with 150 μL of ENST00000430471 knockdown and control HCT116 cells suspensions in the right flanks. The suspensions were at a concentration of 2x106cells/mL. Tumor sizes were measured using a slide caliper after every 3 days. Their volumes were calculated using the following formula: volume (mm3) = 1/2 x length x width2. Ten mice were sacrificed by cervical dislocation after 21 days. The xenografts were subsequently removed after cardiorespiratory arrest.
Cell Invasion Assay
The cell invasion assay was performed in 6.5 mm Transwell chambers (8-μm pore size; EMD Millipore; Merck KGaA). 2 x105 cells were seeded onto the upper chamber coated with Matrigel. The upper chamber contained 200 μL of serum-free medium, while the lower one contained 800 μL of medium supplemented with 20% FBS. The cells were incubated for 24 hours followed by fixing and staining of those on the filter surface. The stained cells were then visualized using a phase-contrast inverted microscope at a magnification of X400. The assay was done in triplicate.
Wound Healing Assay
The wound-healing assay was performed to detect the migration ability of the cells. Cells in the logarithmic growth phase were seeded in 24-well plates and cultured for 24 hours. A 200 μL micropipette tip was then used to create a wound within the cell monolayer. The wound was then visualized and photographed using a Leica DMI3000B microscope after 48 hours. The Image J software was then used to analyze the micrographs of the wounded area. The assay was done in triplicate.
RNA Pull-Down Assay and Mass Spectrometry (MS)
LncRNA ENST00000430471 sequence was cloned to the Nhe I/Sal I site of pCI-neo. The recombinant plasmid was linearized by Sal I to obtain the sense RNA strand. The antisense RNA strand was obtained by linearizing the recombinant plasmid with Nhe I. The biotin-labeled sense RNA (antisense RNA) of ENST00000430471 was transcribed in vitro using Biotin RNA Labeling Mix (Roche Molecular Diagnostics) and T7 (T3) RNA polymerase (New England BioLabs). It was then purified using the EZNA RNA Probe Purification Kit (Omega Bio-Tek, Inc.). The protein lysate from 1x107 HCT116 cells was then incubated with 2 µg of biotinylated RNA and mixed with T1 beads (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA-bound proteins were retrieved from the beads after washing five times in RIPA buffer. The proteins were then heated in SDS buffer and separated using SDS-PAGE. The resultant bands were then silver stained. The bands of interest were then excised and detected using MS analysis.
Statistical Analysis
The GraphPad Prism v6.0 was used for all statistical analysis. Data were presented as mean ± standard deviation from at least three independent experiments. Differences between the two groups were analyzed using the Student’s t-test. One-way ANOVA was used to compare the differences between more than two groups. P values less than 0.05 indicated that there were significant differences between the two groups.
Results
Silencing of ENST00000430471 Inhibits Cell Proliferation Both in vivo and in vitro
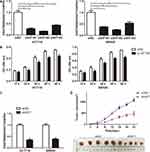
Specific siRNAs (si0471#1, 2 and 3) and shRNA (sh0471) were transfected into HCT116 and SW620 cells to silence the expression of ENST00000430471. This was done to determine whether ENST00000430471 played an important role in CRC proliferation. The transfection efficiencies were confirmed using RT-qPCR (Figure 1A and C; P<0.05). Si0471#2 was found to have the optimal interference efficiency. As such, cells transfected with si0471#2 were used for subsequent experiments. Silencing of ENST00000430471 suppressed cell proliferation in both HCT116 and SW620 cells (Figure 1B; P<0.05). Tumor volumes in mice injected with HCT116-sh0471 cells were significantly smaller compared to those in mice injected with HCT116-shNC cells (Figure 1D). These results strongly suggested that silencing of ENST00000430471 inhibits CRC cell proliferation both in vitro and in vivo.
Silencing of ENST00000430471 Suppresses CRC Cells’ Migration and Invasion
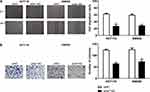
The wound healing assays and Matrigel assays were performed to determine the role of ENST00000430471 in migration and invasion of CRC cells, respectively. Silencing of ENST00000430471 significantly suppressed the migration and invasive ability of both HCT116 and SW620 cells (Figure 2A and B; P<0.05).
ENST00000430471 Interacts with YBX-1
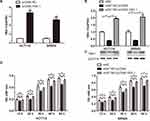
Several studies have postulated that lncRNAs function by binding to a partner protein. Cognizant to this, the RNA pull-down assay with biotinylated ENST00000430471 was performed to identify the proteins interacting with ENST00000430471. This was followed by MS analysis (Figure 3A). Proteins detected included YBX-1, EEF1A1P5, PA2G4 and CCT2 (Table S1). Among the primary proteins, YBX-1 had the highest score from the MS analysis. Moreover, it plays an important role in CRC development. It was therefore selected for subsequent experiments. The subsequent experiments revealed that silencing of ENST00000430471 significantly reduced the expression levels of both YBX-1 mRNA and proteins (Figure 3B and C; P<0.05). Previous studies postulate that lncRNAs either interact with YBX-1 or stabilize the YBX1 protein thereby enabling it to perform its biological functions.12,13 Results herein suggested that there was a pathway that regulated YBX-1 protein transcription when it interacted with ENST00000430471.
Overexpression of YBX-1 Partially Reversed the Tumor Suppression Effects Caused by ENST00000430471 Silencing
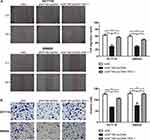
Rescue experiments were performed to validate the role of YBX-1 in ENST00000430471-mediated CRC progression. In the experimental setup, HCT116 and SW620 cells were infected with plasmids containing the YBX-1 expression vector (Figure 4A). The reduction in YBX-1 mRNA and protein expression was restored by pcDNA-YBX-1 in ENST00000430471 silenced HCT116-si0471#2 and SW620-si0471#2 cells (Figure 4B and C). The CCK-8 assay further revealed that the decrease in cell proliferation caused by ENST00000430471 silencing was partially reversed by overexpression of YBX-1 (Figure 4D; P<0.05). Furthermore, overexpression of YBX-1 partially reversed the migration and invasive inhibitory effect induced ENST00000430471 silencing (Figure 5A and B; P<0.05). These results strongly suggested that ENST00000430471 mediated CRC progression partially through regulation of YBX-1 protein expression.
Discussion
Research-based evidences have revealed that dysregulation of lncRNAs promotes the development and metastasis of CRC cells. For example, high MALAT1 expression enhances CRC growth and metastasis.14 In the same line, MIR503HG is downregulated in CRC and contributes to CRC migration and invasion.15 However, further studies are needed to identify additional functions of lncRNAs in CRC development.
Previous studies postulate that ENST00000430471 is highly expressed in CRC and further promotes its development.11 Herein, silencing of ENST00000430471 suppressed proliferation, migration and invasion of CRC cells in vitro. HCT116 was selected for xenograft experiment because its tumor volume was larger than that of SW620 in vivo.16 Silencing of ENST00000430471 reduced the tumor volume in vivo. These results indicated that ENST00000430471 acted as an oncogene in CRC. Cognizant to this, further studies are required to explore the underlying mechanism of ENST00000430471.
LncRNAs regulate tumor biological functions through multiple pathways.17 They exert their biological function by binding to a partner protein thus affecting the expression of target genes.18 For instance, LncRNA OCC-1 regulates the levels of many mRNAs at the post-transcriptional level by binding to and destabilizing HuR.19 In the same line, LncRNA SNHG1 regulates CRC cell growth by interacting with polycomb repressive complex 2 thereby modulating the histone methylation promoter of Kruppel like factor 2 and cyclin-dependent kinase inhibitor 2B.20 Herein, the RNA pull-down assay and subsequent MS analytical technique revealed that ENST00000430471 interacted with YBX-1, EEF1A1P5, PA2G4 and CCT2 proteins. YBX-1 had the highest score and was thus selected for subsequent experiments.
YBX-1 is an oncogene. High expression of YBX-1 is associated with poor prognosis and local recurrence in CRC.21,22 Overexpression of YBX-1 promotes proliferation, migration and induces oxaliplatin resistance in CRC cells.23,24 Silencing of YBX-1 inhibits the malignant progression of CRC cells by reversing epithelial‑mesenchymal transition.25 Several studies have postulated that lncRNAs such as HULC, MIR22HG, HOXC-AS3, GAS5, and TP53TG1 can bind to YBX-1. The lncRNAs regulate phosphorylation, protein turnover, stability and nuclear localization of YBX-1.12,26-29 Herein, ENST00000430471 silencing reduced YBX-1 expression both at the mRNA and protein level. This suggested that another pathway regulates YBX-1 mRNA expression while it interacts with ENST00000430471. Moreover, overexpression of YBX-1 partially attenuated the suppression of cell proliferation, invasion and migration induced by ENST00000430471 silencing.
Conclusion
Silencing of ENST00000430471 inhibits proliferation, migration, and invasion of CRC cells by regulating YBX-1 expression. These results provide baseline information that is essential in the identification of effective therapeutic targets for CRC therapy.
Ethics Statement
All animal-based experiments were approved by the Animal Care and Use Committee of Xishan People’s Hospital (Jiangsu, China). They also followed the ARRIVE guidelines.
Acknowledgments
This study was financially supported by grants from the Medical and Public Health Technology Research and Development Foundation of Wuxi (Grant No. CSE31N1726) and Natural Science Foundation of Wuxi Youth (Grant No. Q201745).
Disclosure
The authors report no conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
3. Ribatti D, Mangialardi G, Vacca A. Stephen paget and the ‘seed and soil’ theory of metastatic dissemination. Clin Exp Med. 2006;6(4):145–149. doi:10.1007/s10238-006-0117-4
4. Lan Y-T, Yang S-H, Lin J-K, et al. Genetic variations are associated with lymph node metastasis in colorectal cancer patients. J Surg Oncol. 2014;110(3):307–312. doi:10.1002/jso.23613
5. Sun W, Shi Y, Wang Z, et al. Interaction of long-chain non-coding RNAs and important signaling pathways on human cancers (review). Int J Oncol. 2018;53(6):2343–2355. doi:10.3892/ijo.2018.4575
6. Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi:10.1186/s12943-016-0530-6
7. He X, Tan X, Wang X, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35(12):12181–12188. doi:10.1007/s13277-014-2526-4
8. Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. doi:10.1158/0008-5472.CAN-11-1021
9. Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23(9):1446–1461. doi:10.1101/gr.152942.112
10. Wang M, Han D, Yuan Z, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9(12):1149. doi:10.1038/s41419-018-1187-4
11. Yang P, Xu Z-P, Chen T, et al. Long noncoding RNA expression profile analysis of colorectal cancer and metastatic lymph node based on microarray data. Onco Targets Ther. 2016;9:2465–2478. doi:10.2147/OTT.S102348
12. Su W, Feng S, Chen X, et al. Silencing of long noncoding RNA MIR22HG triggers cell survival/death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res. 2018;78(12):3207–3219. doi:10.1158/0008-5472.CAN-18-0222
13. Zhang W, Du M, Wang T, et al. Long non-coding RNA LINC01133 mediates nasopharyngeal carcinoma tumorigenesis by binding to YBX1. Am J Cancer Res. 2019;9:779–790.
14. Wu S, Sun H, Wang Y, et al. MALAT1 rs664589 polymorphism inhibits binding to miR-194-5p, contributing to colorectal cancer risk, growth, and metastasis. Cancer Res. 2019;79(20):5432–5441. doi:10.1158/0008-5472.CAN-19-0773
15. Chuo D, Liu F, Chen Y, et al. LncRNA MIR503HG is downregulated in Han Chinese with colorectal cancer and inhibits cell migration and invasion mediated by TGF-β2. Gene. 2019;713:143960. doi:10.1016/j.gene.2019.143960
16. Fang T, Liang L, Hu WJ, et al. Establishment and comparison of subcutaneous xenograft models of three human colorectal cancer cell lines in nude mice. Thirteen Symp Lab Anim Sci East China. 2015;23–27.
17. Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74. doi:10.1007/978-981-10-5203-3_2
18. Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17:106–116. doi:10.1093/bib/bbv031
19. Lan Y, Xiao X, He Z, et al. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46:5809–5821. doi:10.1093/nar/gky214
20. Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17(1):141. doi:10.1186/s12943-018-0894-x
21. Yan X, Yan L, Zhou J, et al. High expression of Y-box-binding protein 1 is associated with local recurrence and predicts poor outcome in patients with colorectal cancer. Int J Clin Exp Pathol. 2014;7(12):8715–8723.
22. Shiraiwa S, Kinugasa T, Kawahara A, et al. Nuclear Y-box-binding protein-1 expression predicts poor clinical outcome in stage III colorectal cancer. Anticancer Res. 2016;36:3781–3788.
23. Nagasu S, Sudo T, Kinugasa T, et al. Y‑box‑binding protein 1 inhibits apoptosis and upregulates EGFR in colon cancer. Oncol Rep. 2019;41(5):2889–2896. doi:10.3892/or.2019.7038
24. Tsofack SP, Garand C, Sereduk C, et al. NONO and RALY proteins are required for YB-1 oxaliplatin induced resistance in colon adenocarcinoma cell lines. Mol Cancer. 2011;10(1):145. doi:10.1186/1476-4598-10-145
25. Yan XB, Zhu QC, Chen HQ, et al. Knockdown of Y-box binding protein1 inhibits the malignant progression of HT29 colorectal adenocarcinoma cells by reversing epithelial-mesenchymal transition. Mol Med Rep. 2014;10:2720–2728. doi:10.3892/mmr.2014.2545
26. Liu Y, Zhao J, Zhang W, et al. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159. doi:10.1038/srep10159
27. Diaz-Lagares A, Crujeiras AB, Lopez-Serra P, et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc Natl Acad Sci U S A. 2016;113(47):E7535–E7544. doi:10.1073/pnas.1608585113
28. Li D, Liu X, Zhou J, et al. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2017;65(5):1612–1627. doi:10.1002/hep.29010
29. Zhang E, He X, Zhang C, et al. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19(1):154. doi:10.1186/s13059-018-1523-0
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.