Back to Journals » International Journal of Nanomedicine » Volume 17
Enhancing Cutaneous Wound Healing Based on Human Induced Neural Stem Cell-derived Exosomes
Authors Li J , Gao H, Xiong Y, Wang L, Zhang H, He F, Zhao J, Liu S, Gao L, Guo Y, Deng W
Received 4 July 2022
Accepted for publication 23 November 2022
Published 5 December 2022 Volume 2022:17 Pages 5991—6006
DOI https://doi.org/10.2147/IJN.S377502
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Jing Li,* Hong Gao,* Yue Xiong,* Ling Wang, Haojie Zhang, Fumei He, Jingxin Zhao, Shuna Liu, Liqian Gao, Ying Guo, Wenbin Deng
School of Pharmaceutical Sciences (Shenzhen), Shenzhen Campus of Sun Yat-Sen University, Shenzhen, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ying Guo; Wenbin Deng, Email [email protected]; [email protected]
Background: Wound healing of skin is a complicated process. Cutaneous innervation and neurotrophic factors could participate in multiple stages of wound healing. Neurotrophic factors are mainly produced and released by neurons and neural stem cells (NSCs) which could be obtained in large quantities from human-induced pluripotent stem cells (iPSCs) in vitro. However, the potential wound healing effects of NSC secretions, such as exosomes, are unexplored yet.
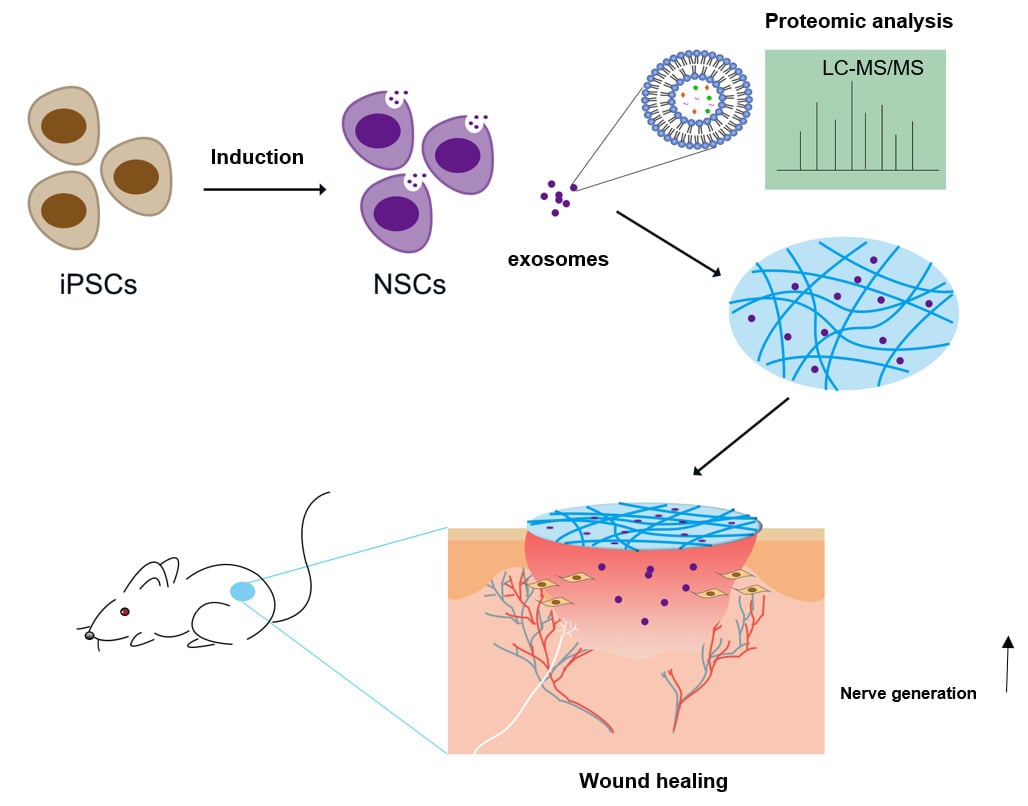
Methods: NSCs-derived exosomes (NSC-exo) and iPSCs-derived exosomes (iPSC-exo) were isolated from the cell culture supernatants by centrifugation, and then quantified and characterized. The effects of these exosomes on the migration of human dermal fibroblasts (HDF) cells and the tube formation of human umbilical vein endothelial cells (HUVECs) were investigated in vitro. And the in vivo wound healing effect of these exosomes were tested on the mouse skin trauma model. Therefore, a dipeptide/hyaluronic acid (Nap-FF/HA) composite hydrogel was used to encapsulate the exosomes as a sustained release carrier. Histological observations were performed to evaluate the wound healing effect of exosomes. Furthermore, the non-labeling proteomic analysis was performed to explore the possible mechanisms of NSC-exo on wound healing.
Results: We obtained extracellular vesicles in a bowl-like structure with membranes which meet the general standards of exosomes. NSC-exo showed promotion effect on the migration of HDF cells and the tube formation of HUVECs in vitro. In a mouse skin injury model, NSC-exo enhanced the wound healing and the Nap-FF/HA hydrogel that contained exosomes did so with less drug frequency by sustaining release of exosomes. Further proteomic analysis demonstrated that the carried neurotrophic factors and immunity-related proteins in NSC-exo may play a functional role in wound healing.
Conclusion: NSC-exo may enhance wound healing via neurotrophic factors and immunomodulation.
Keywords: wound healing, neural stem cells, exosomes, proteomic analysis
Graphical Abstract:
Introduction
Skin wound healing is a multilayered complicated process which mainly includes four overlapped stages: hemostasis, inflammation, proliferation, and remodeling.1 Damaged skin cells release a large number of debris and active substances which could induce inflammation response. Excessive and persistent inflammation hinders the healing process and finally leads to chronic wounds. Given the that natural ability of epidermal tissue regeneration is not enough for curing severely injured patients,2–4 more effective therapies need to be explored.
In fact, the skin has been considered as neuroimmunoendocrine organ, and the neural control of skin function has been explicitly reviewed. The skin is mainly composed of three layers: epidermis, dermis, and hypodermis. These nerves are distributed in hypodermis, but all layers contain abundant nerve endings.5 Emerging evidences showed that cutaneous innervation may play important roles in wound healing.6,7 Numerous neuropeptides secreted by the sensory and autonomic nerve fibers play essential roles during the distinct phases of wound healing.6 In addition, the skin produces neuromediators and neurotrophic factors that target nerve fibers, thereby modulating inflammation, immune responses during host defense, pain, and pruritus.8
Various studies have reported that using antioxidant agents or immunosuppressants could mediate the inflammation response, promoting wound healing.9,10 However, emerging studies of stem-cell based therapies provide prospects on tissue regenerative. Cell therapies, such as bone marrow-derived mesenchymal stem cells (MSCs), induced pluripotent stem cells (iPSCs) and fibroblast lineages have been applied on different wounds models in various studies.11,12 Induced pluripotent stem cell technology has opened up unique opportunities for developing human brain cells such as expandable neural stem cells (NSCs) and neurons which were hard to obtain in the past. Human NSCs possessed neural differentiation potency and were once employed as neuronal components of human skin equivalent.5 NSCs were reported that played a critical role in the processes of wound healing of spinal cord injury.13 However the potential wound healing effects of NSC secretions are unexplored.
Recently, the exosomes derived from specific cells exert promising therapeutic effects on the wound healing. Compared with stem cell therapy, the exosomes, which comprise nanosized membrane complexes secreted by cells as cargos for cell-cell communication, represent an attractive alternative approach.14,15 In addition, exosomes have advantages such as good biodistribution and do not form teratoma-like tumor masses. Extracellular vehicles derived from mouse neural stem cells were found to have therapeutic potential in brain ischemic injury and spinal cord injury.16–18 Recently, the anti-inflammatory and neurogenic properties of extracellular vesicles from human iPSC-derived NSCs have been studied via analysis of miRNA and protein signatures.19 The influence on wound healing remains unknown. Therefore, we put forward a hypothesis that the exosomes of the human iPSCs derived NSCs (NSC-exo) had promoted effect on wound healing.
In this study, the NSC-exo were collected and administrated to cutaneous wound healing mouse model. Interestingly, we found that the NSC-exo had promoted effects on the migration of human dermal fibroblasts (HDF) cells and the tube formation of human umbilical vein endothelial cells (HUVECs). We firstly reported that the effects of NSC-exo promoted wound healing. In addition, the combination of a dipeptide/hyaluronic acid (Nap-FF/HA) composite hydrogel and NSC-exo could achieve similar effects on wound healing with less administration frequency. Furthermore, the results of non-label proteomic analysis indicated that the NSC-exo carried high content of neurotrophic factors such as neuron-derived neurotrophic factor (NDNF), immunoregulation proteins, and other factors related to wound healing, which may provide the possible repair mechanism. In other words, this work revealed a novel role of NSC-exo on wound healing, which might contribute to the development of novel therapies for skin injury in the future.
Materials and Methods
Cell Culture and Differentiation
Human urine cell-derived iPSC cell line, UE017C1, was kindly provided by Professor Kaijie Chen from Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences (Guangzhou, China), and cultured on Matrigel®-coated six-well plates (Corning, USA) in mTeSR™ medium (Stem Cell Technologies, Canada) and passaged by using ReLeSR™ (Stem Cell Technologies, Canada). The generation of NSCs from iPSCs was according to the previous protocol as described.20–22 Briefly, the primitive NSCs were induced when iPSCs at 20% confluence. The iPSC culture medium was replaced with a neural induction media (NIM) supplemented with 4 μM CHIR99021 (MCE, USA) and 3 μM SB431542 (MCE, USA), 0.1 μM Compound E (MCE, USA), for seven days. NIM was prepared by mixing Advanced DMEM/F-12 (Gibco, USA) and neurobasal media (Gibco, USA) in a 1:1 ratio, supplemented with N2 (1×, Stem Cell Technologies, Canada), B27 (1×, Gibco, USA), Glutamax (1%, Gibco, USA), hLIF (10 ng/mL, PeproTech, USA). Dorsomorphin (2 µM; Sigma-Aldrich, USA) was added to the culture for two days and excluded for another five days. The culture was then dissociated using Accutase™ solution (Millipore, USA) at a dilution of 1:3, cultured in neural induction media supplemented with 3 μM CHIR99021 and 2 μM SB431542, 5 μM Y-27632 (ROCK inhibitor; MCE, USA) on Matrigel®-coated plates. Followed six passages, the cells were passaged at dilution of 1:4 regularly. The culture medium was replaced with fresh media every other day. The NSCs from different passages were confirmed through immunofluorescence staining and RT-PCR for Nestin, Pax6, and Sox2. The use of UE017C1 iPSCs cell lines had ethical or institutional review board approval.
Immunofluorescence
NSCs were washed with PBS for three times and fixed in 4% paraformaldehyde for 15 min. The fixed cells were washed with PBS and penetrated with 0.2% Triton X-100 for 20 min. Then the cells were washed with PBS and incubated in normal goat serum (Bosterbio, USA) for 40 min at room temperature. The cells were then incubated with mouse anti-Nestin (Cell Signaling Technology, USA), rabbit anti-Pax6 (Cell Signaling Technology, USA), and rabbit anti-Sox2 (Cell Signaling Technology, USA) overnight at 4°C. On the next day, cells were washed with PBS containing 0.1% Tween 20 (PBST) (BBI Life Science, Shanghai, China) and incubated with Alexa Fluor-conjugated secondary antibodies (1000×, Invitrogen, USA) in PBST for one hour at room temperature (RT). Nuclei were stained with DAPI. Images were captured by a FV3000 confocal laser scanning microscopy (Olympus Optical Co. Ltd, Tokyo, Japan).
RNA Extraction and RT-PCR Analysis
To identify the differentiated neural stem cells with iPSCs from gene level, real-time PCR was performed. Total RNA was extracted using the RNA-Quick Purification Kit (Exunbio, YiShan Biotech, Shanghai, China). Reverse transcription was performed with 1 μg RNA using EasyScript All-in-One First-Strand cDNA Synthesis SuperMix for qRCR (TransGen Biotech, Beijing, China). Real-time PCR was carried out using PerfectStart Green qPCR SuperMix (TransGen Biotech, Beijing, China). The primers for the Nestin, Pax6, β-actin, were listed in Table 1. The expression of gene was normalized to that of β-actin in all samples. After normalization, data were transformed as log10 of the target mRNA signal relative to the untreated control sample.
 |
Table 1 The Sequence of Primers |
Isolation of Exosomes
Exosomes were purified from the supernatants of iPSCs and NSCs. The conditioned medium was collected when replaced with fresh media. The harvest of NSCs exosomes was performed by ultracentrifugation as described.23 Briefly, the collected medium was first centrifuged for 10 min at 600 g and then 20 min at 2000 g to remove cells and large cell debris. The supernatant was then centrifuged for 40 min at 10,000 g, and the resulting supernatant passed through a 0.22 μm filter to remove residual cell debris. After then, the filtered supernatant was ultracentrifuged at 100,000 g for one hour, the pellet was washed with PBS and ultracentrifuged at 100,000 g for another one hour. The pelleted exosomes were resuspended in 100 μL of PBS, quantified by Pierce™ BCA protein assay kit, and preserved at −80°C for later analysis. All centrifuge process were performed at 4°C.
Dynamic Light Scattering (DLS)
The size distribution and zeta potential of exosomes were measured by an instrument (NanoBrook 90Plus PALS, Brookhaven Instruments, USA). Exosomes were diluted with PBS at a ratio of 1:200.
Western Blotting
The protein concentrations of collected exosomes were measured by BCA protein assay (Thermo Scientific, USA). Equal amounts of protein were separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Bio-Rad, USA). Membranes were blocked with 5% non-fat milk (BD Biosciences, USA) for one hour and incubated with primary antibodies overnight at 4°C After washing with 1×TBST, membranes were incubated with appropriate secondary antibodies at room temperature for two hours, and visualized by using the HRP-enhanced chemiluminescence substrate (Bio-Rad). The primary antibodies, anti-ALIX, anti-CD63, anti-TSG101 and anti-GAPDH, were purchase from Abcam (UK). The secondary antibodies were goat anti-rabbit (Abcam) and goat anti-mouse (Abcam).
Scratch Assay
To explore the influence of NSC-derived exosomes on the migration of skin cells, a scratch assay was performed. HDF cells were cultured with DMEM/F12 supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C under 5% CO2 and pre-seeded in 48-well plates. At 80% confluence, cells were stained with Hoechst 33,342 and AM-calcium, then scratched using 10 μL pipette tip and washed with PBS. Cells were incubated with exosomes (100 μg/mL) from iPSCs, NSCs and bFGF (10 ng/mL) for 24 h. The control group was treated with DMEM. Images of scratch areas were taken at 0, 12, and 24 h time points. The migration percentage was measured by ImageJ software.
Tube Formation
To investigate the role of the NSC-exo in angiogenesis in vitro, a tube formation assay on HUVECs which is a crucial step in angiogenesis was performed. HUVECs were cultured with DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C under 5% CO2. Matrigel® was pre-thawed in freezer at 4°C. Thirty microliters of undiluted Matrigel® was added to wells of 96 well plates uniformly via precooling tips. Then the plates were incubated at 37°C for 30 min. HUVECs were digested by trypsin, resuspended in DMEM supplemented with 2% FBS, and seeded on the wells of Matrigel®-coated plates at a density of 2×105 cells/mL. 20 μg/mL iPSC-exo and NSC-exo were added to corresponding wells respectively. The control group was treated with DMEM supplemented with 2% FBS. Each group was provided with three repeated wells. The plates were put into a cell incubator. After six hours, images of tube formation were taken using a Nikon Eclipse Ti2-U microscope and analyzed by ImageJ software.
Hydrogel Preparation
The Nap-FF/HA composite hydrogel (0.2 wt %) was prepared by one-pot reaction. The Nap-FF peptide powder (GL Biochem, Shanghai, China) was dissolved in DMSO (Sigma-Aldrich, St Louis, MO, USA) to 100.0 mg/mL. The hyaluronic acid (HA) powder (AR, MW: 800–1000 KDa, Heowns Biochem, Tianjin, China) was dissolved in PBS (Gibco) to 10.0 mg/mL. The peptide solution was mixed with HA solution at a ratio of 1:4 (wt/wt), shaken for 10 s and left to form hydrogel. For the preparation of exosomes-loaded hydrogels, exosomes (final concentration at 1.0 μg/μL) were mixed with HA firstly, then mixed with Nap-FF and left until hydrogel formed. The formation of hydrogels was confirmed by inverting the reaction tubes.
Rheology Experiment
Rheological measurements were carried out on an ARES/RFS high rotational rheometer (TA Instruments) with a 25 mm paralleled plate at 25°C. Briefly, 500 μL of 0.2 wt % Nap-FF/HA hydrogel was sandwiched between the cone plates followed by rheological measurements. The storage modulus G′ and loss modulus G″ were obtained as function curves of frequency. Dynamic oscillatory frequency sweep measurements were conducted at a 0.5% strain amplitude with the angular frequency ranging from 0.1 to 100 rad/s.
Transmission Electron Microscopy (TEM)
The morphology of isolated exosomes and obtained hydrogel were observed by TEM. For the sample preparation, 10 μL purified exosomes which suspended in PBS and droplets of diluted hydrogel solution were placed on formvarcarbon-coated copper grids, respectively, left for 5 min. Then the samples were removed from the edge of the grids with filter paper. Three percent phosphotungstic acid solution were added to the grids and stained for 5 min. Grids were washed with ultrapure water for 5 min and left to dry for overnight. Exosomes and hydrogel were imaged by TEM (HC-1, Hitachi) at 80 kV.
Scanning Electron Microscopy (SEM)
To characterize the surface morphology of the Nap-FF/HA hydrogels, scanning electron microscope images were captured using a JSM-6330F SEM (JEOL, Japan). The hydrogel samples were prepared by lyophilization overnight in an Alpha 1–4LD plus freeze drier (Mettler Toledo, Switzerland). Then the samples were placed on the working stage using black double-sided adhesive tape. The surface of the hydrogel samples was sputter-coated with gold for seconds under argon before imaging.
In vivo Wound Healing Experiments
Balb/c mice (male, aged 4–6 weeks, weight 18–22 g) were purchased from the Laboratory Animal Center of Sun Yat-sen University (Guangzhou, China). All the mice were handled in accordance with the protocol approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (Approval Number: SYSU-IACUC-2022-001111) and followed Laboratory Animal-Guidelines for ethical review of animal welfare (Standard Number: GB/T 35892–2018). Following a week of acclimatization, animals were intraperitoneally injected with 1% (w/v) pentobarbital sodium (Sigma-Aldrich, USA) at a dose of 50 mg/kg, the hair on the back was removed with hair removal creams. A circle of full-thickness skin wound measuring 10×10 mm2 (≈100 mm2) was created on the dorsal middle region of the mice. The mice were grouped and applied with 50 μL PBS, NSC-exo (1.0 μg/μL), iPSC-exo, NapFF/HA hydrogel, NapFF/HA/NSC-exo, NapFF/HA/iPSC-exo. Then wound edges were fixed by silicone loops and protected by wrapping 3M Tegaderm. The groups of exosomes were only treated daily for a week. The groups of hydrogels containing exosomes were treated every three days for three times. All wounds were photographed every day. The wound healing rates were measured by ImageJ software.
Histology and Immunohistochemistry Analysis
Animals were sacrificed on postoperative day 12, the damaged skins were excised and fixed in 4% paraformaldehyde overnight. The fixed skins were embedded in paraffin, cut into slices for H&E and Masson’s staining according to universal protocols. For immunohistochemistry analysis, the skin slices were rehydrated, incubated in citrate antigen retrieval solution, blocked with 3% bovine serum albumin, stained using primary antibodies to alpha smooth muscle (α-SMA; Sabbiotech, USA), CD31 (Service Bio, Wuhan, China) and the neural regeneration marker growth associated protein 43 (gap43; Cell Signaling Technology, USA) and TUJ1 (Cell Signaling Technology, USA). After being incubated with appropriate secondary antibodies, the slides were reacted with DAB substrate solution (Service Bio, Wuhan, China) and counterstained with hematoxylin, mounted with resin. The images of the slides were captured by Nikon Eclipse Ti2-U microscope, and analyzed by ImageJ software.
Proteomic Analysis of NSC-exo
Proteomic samples of exosomes derived from NSCs were prepared by in-gel digestion as described.24 Briefly, equal amounts of exosomes (20 μg) and cell lysates of neural stem cells were resolved by 10% SDS-PAGE. The protein bands were cut into cubes (1×1 mm) and transferred into microcentrifuge tubes. The protein cubes were destained using 50 mM ammonium bicarbonate/acetonitrile (1:1, v/v), in-gel reduced by 10 mM DTT (Bio Froxx, Guangzhou, China) and alkylated by 55 mM iodoacetamide (IAA, Sigma, USA) in the dark. Followed in-gel digestion with 1 μg trypsin (Promega, USA) at 37°C overnight, 0.1% formic acid (Fisher Scientific) was added to stop the digestion. Peptide fragments were extracted by 0.1% formic acid/acetonitrile and desalted using Pierce® C18 Spin Columns (Thermo Fisher, USA). The final purified peptides were dissolved in 0.1% formic acid for further LC-MS/MS analysis using Nano LC-Q Exactive Plus (Thermo Fisher). The injection volume was 10 μL, the flow rate was 500 nL/min. The raw data were processed using Proteome Discoverer (PD) 2.1 software (Thermo Scientific), the MS/MS spectra were searched against the human proteome (UniProt). The peptide and protein identifications were filtered by using PD to control the false discovery rate (FDR) <5%.
Gene ontology (GO) analysis was performed using the DAVID bioinformatics tool.25 The pathway analysis was performed using Panther database. Protein interaction network analysis was performed using STRING online tool.
Statistical Analysis
All quantitative data were analyzed with GraphPad Prism 5.0 software (GraphPad Software Inc., San Diego, CA, USA), and shown as mean ± standard deviations (SD). The significances were analyzed by using the Student’s t-test for two groups comparisons and one way ANOVA with post hoc for multiple groups comparisons. And statistically significance was determined at P<0.05.
Results and Discussion
hiPSCs-derived NSCs Differentiation and Exosomes Characterization
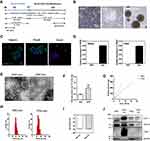
NSCs were obtained from iPSCs according to previous reports following the schedule presented in Figure 1A, by sequentially treating small molecule inhibitors.20–22 As shown in Figure 1B, the cell morphology of iPSCs and NSCs were quite different, and the cluster of NSCs showed more density with scattered cells around. If cultured on low-attachment plates, NSCs grew and formed neurospheres of different sizes, confirming the spheroid-forming properties of NSCs. The biomarkers of NSCs were characterized via immunofluorescence. As shown in Figure 1C, the NSCs showed high expression of Nestin, Pax6 and Sox2. The qPCR results indicated that the Nestin and Pax6 were high-expressed in NSCs (Figure 1D). The obtained induced NSCs were collected with high purity for later experiments. The iPSCs-derived and NSCs-derived exosomes were isolated from the cell culture supernatants by centrifugation. And the obtained exosomes were quantified and characterized (Figure 1E–J). The morphology of obtained exosomes was imaged by transmission electron microscope (TEM), the images showed a bowl-like structure with membranes and elliptic or circular shapes, and the size of iPSC-exo was larger than NSC-exo (Figure 1E). The exosomes protein acquired from iPSCs was about 0.4 μg per 106 cells, which was twice as much as that from NSCs (Figure 1F). It showed that the same cell number of NSCs secrete fewer exosomes than iPSCs. Besides, a linear relationship between the number of cells and the yields of exosomes was illustrated (Figure 1G), which indicated a stable production of exosomes during cell culture. The results of dynamic light scattering (DLS) in Figure 1H indicated that the size distribution of NSC-exo (36.36~56.51 nm) was smaller than iPSC-exo (50.75~105.7 nm), which was corresponded with the TEM results. Furthermore, both of the exosomes showed negative zeta potential (Figure 1I) and expressed the biomarkers of exosomes from stem cells: ALIX, TSG101, and CD63 (Figure 1J). These results demonstrated that the isolated exosomes of iPSCs or NSCs meet the general standards of the definition of exosomes.
NSC-exo Promoted the Migration of HDF Cells and Tube Formation of HUVECs in vitro
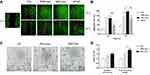
The scratch assay of skin cells is one of the models to simulate skin trauma in vitro. In order to explore the effect of NSC-exo on skin trauma in vitro, the effect of NSC-exo on HDF cell migration was first explored via scratch assay. Previous studies have shown that iPSC-exo could promote cell migration,26 and while bFGF applied to wounds could promote fibroblast migration.27 Therefore, iPSC-exo and bFGF were used as positive controls. As shown in Figure 2A and B, the migration rate in the NSC-exo group was up to nearly 70% in 12 h, whereas there were only 20% in negative control group and 40% in bFGF group. There was significant migration compared with the control group (P<0.001, vs Control) and bFGF group (P<0.01, vs bFGF). The migration rate of iPSC-exo group was about 60%, but there was no significant difference between iPSC-exo and NSC-exo group. After 24 h, the group treated with NSC-exo almost completely migrated (the migration rate was about 97%, P<0.001, vs Control), which was quite different from iPSC-exo group (P<0.05, vs iPSC-exo). These results demonstrated for the first time that the obtained NSC-exo could significantly promote the migration of HDF cells, which also preliminarily demonstrated the potential of NSC-exo on wound healing.
Angiogenesis is indispensable for successful wound healing.28 The process of angiogenesis mainly includes cell proliferation, migration and tube formation. The tube formation assay is a rapid and quantitative method to study angiogenesis.29 To investigate the angiogenesis ability of NSC-exo, the tube formation experiment on human umbilical vein endothelial cells (HUVECs) was performed. HUVEC is the most suitable cell for simulating angiogenesis in vitro. It possesses potentials of stem cells and property of forming tubular structure in extracellular matrix. Generally, HUVECs are adherent cultured on ordinary cell culture plates. Using Matrigel® to simulate the components of extracellular matrix and seeding a certain number of cells on the gel, the cells can form a tubular structure within 24 h. The effects of different drugs on cell angiogenesis can be accessed by analyzing the nodes, branches, or vessel length of the tubular structure.
As shown in Figure 2C and D, HUVECs were seeded in 96-well plates which were pre-coated with Matrigel® at a density of 2×105 cells/mL, and treated with 20 μg/mL iPSC-exo or NSC-exo. After six hours, a cross-linking tubular structure was formed in both exosomes’ groups, which was significantly better than that in the control group. The quantitative analysis showed that the number of branches in NSC-exo group was about 1.5 times that of the control group (P<0.01, vs Control), and the total branching length was about 1.9 times that of the control group (P<0.001, vs Control). The results indicated the good ability of NSC-exo to promote tube formation of HUVECs. However, the number of branches in iPSC-exo group was about 1.3 times that of the control group (P<0.05, vs Control), and the total branching length was about 1.5 times that of the control group (P<0.01, vs Control), in which the total branching length was different from that of NSC-exo group (P<0.05, vs NSC-exo). The results demonstrated that the angiogenesis ability of NSC-exo was kind of looked better than that of iPSC-exo.
Nap-FF/HA Hydrogel Preparation and Characterization
The Nap-FF/HA (N/HA) hydrogel was designed and reported by our recent work.30 The chemical structures of Nap-FF and HA and the images of formed hydrogels were exhibited in Figure 3A. A mixture of peptide solution and hyaluronic acid was mixed in a small glass bottle. After stewing at RT, the glass bottle was inverted, and the mixture had been fixed to the bottom of the bottle, indicating that a transparent and homogeneous hydrogel has been successfully formed.
Next, rheology experiments were used to measure the mechanical properties and the state of hydrogel. The elastic modulus G′ represents how much energy the material stores through elasticity, also known as storage modulus, the viscosity modulus G″ indicates the ability of a material to dissipate energy, also known as loss modulus. As shown in Figure 3B, the elastic modulus and viscosity modulus of the hydrogel were relatively constant as the frequency changed, indicated that the mechanical properties of hydrogels are independent on frequency, which indirectly proved that the composite hydrogel had relatively stable crosslinking network inside. The elastic modulus of the hydrogel (about 1 kPa) was always greater than that of the viscous modulus (about 100 Pa) during the change of frequency, showed that the hydrogel formed was solid-like state. In order to further characterize the internal structure of hydrogel, the result of scanning electron microscope was shown in Figure 3C. The diameter of the internal micropores of the hydrogel was about 1–4 μm, which indicated that the inner micropore structure of the hydrogel might encapsulate and sustain exosome release on skin wounds. The characterization result of TEM was showed in Figure 3D. The hydrogel was composed of numbers of long nanofibers with a diameter of about 6.72±1.23 nm. These fine fibers were cross-linked into a network structure. Above results confirmed the formation of hydrogels and provided the possibility of which to load exosomes allowed by the inside structure.
NSC-exo Promoted in vivo Wound Healing of Mice
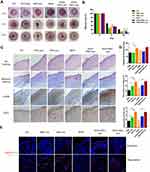
The full-thickness skin injury in mice involves the loss of subcutaneous tissue, sensory nerve endings and some neuroimmune functions. The wound healing process generally takes about two weeks, which can be used as a short-term model for cutaneous wound healing. In order to compare the effects of the free exosomes and the exosome-loaded hydrogels, different frequency of administration was taken. The control group of exosome free group was treated with PBS (Control). In the hydrogel group, blank hydrogel was set as the control group (N/HA). To investigate the sustained release effect, the hydrogel contained exosomes was administered every three days. Results were as shown in Figure 4A and B, for the groups without hydrogels on the fifth day after injury, the wound area of control group was about 50%, and the wound areas of iPSC-exo group and NSC-exo group had shrank to about 30%. There was significant difference between NSC-exo group and control group (P<0.05, vs Control). On the tenth day, the wound in the control group had recovered to about 20%, while both of that in iPSC-exo and NSC-exo groups were about 9%. There was significant difference in both iPSC-exo group (P<0.01, vs Control), and NSC-exo group (P<0.01, vs Control). These results indicated that both exosomes could significantly promote wound healing of mice. Furthermore, NSC-exo group exerted comparative therapeutic effect on wound healing compared with iPSC-exo group.
When the exosomes were loaded into the N/HA hydrogel, the wound area of N/HA group was about 50% on the fifth day after injury. The wound area of N/HA-iPSC-exo group was about 36%, and that of N/HA-NSC-exo group was about 34%. On the tenth day, the wound area of N/HA group was about 25%, that of N/HA-iPSC-exo group was about 12% (P<0.001, vs N/HA), and that of N/HA-NSC-exo group was about 4% (P<0.001, vs N/HA). There was significant difference among different exosomes loaded hydrogels (P<0.05, N/HA-NSC-exo vs N/HA-iPSC-exo). The results demonstrated that hydrogel contained exosomes also significantly promoted wound healing. Interestingly, on the tenth day, the wound healing effect of N/HA-NSC-exo group was better than NSC-exo group even without significance. On the one hand, the above results showed that the exosomes-loaded hydrogels could achieve considerable repair effect via controlled release compared with the groups without hydrogels, which also reduced the drug frequency and improved the bioavailability of exosomes. On the other hand, the effect of N/HA-NSC-exo might be better than that of N/HA-iPSC-exo on wound healing. However, there was no significant difference between the two exosome groups without hydrogels. The reason of the outcome should be further discussed, which might be the different size and composition between two exosomes or that the different loss of exosomes in the experiment. In addition, the introduction of hydrogels might exhibit more stable effects of exosomes by prolonging the stay of exosomes on the wounds.
On the twelfth day after skin trauma, the mice in each group were anesthetized, the wound area was cut, and paraffin sections were made to further evaluate the wound healing. H&E staining was used to evaluate the regeneration of cutaneous tissue structure. For example, re-epithelization is the main process in the proliferation phase on wound healing. The degree of skin recovery in each group can be estimated by epidermal thickness. Moreover, the collagen regeneration in skin can be evaluated by Masson trichrome staining, and the increase of SMA (smooth muscle actin) indicates the healing of skin smooth muscle cells, which helps to pull the contraction of skin wound and accelerate wound healing. In addition, angiogenesis is the main process in the proliferative phase after the inflammatory phase, and CD31 is a platelet endothelial cell adhesion molecule, which can be used to mark the number of new blood vessels and vascular regeneration. The skin sections of each group were treated with H&E staining, Masson staining and immunohistochemistry of CD31 and SMA, followed by the semiquantitative analysis via IHC Toolbox tool in ImageJ software. As shown in Figure 4C and D, the wound healing effect of hydrogels contained exosomes groups were a little bit better than exosomes without hydrogel groups from the comprehensive re-epithelialization thickness, collagen regeneration and expressions of CD31, but there was no significant difference between them. The effect of N/HA-NSC-exo group was superior and the epidermal thickness was increased in NSC-exo group (P<0.01), the collagen distribution and the expression of CD31 were higher than those in groups iPSC-exo and PBS. And the expression of these results indicated that the NSC-exo promoted cutaneous wound healing by re-epithelialization, collagen regeneration, skin contraction and angiogenesis. In addition, the combination of hydrogels also improved the bioavailability and therapeutic effect of exosomes, and the effect on wound healing might be a combined results from the exosomes and hyaluronic acid.
The distribution of subcutaneous nerves includes sensory nerve endings in the skin epithelial layer, receptors in the dermis and oval ring body in the deep dermis. Growth related protein gap43 and neuron specific class III skeleton protein TUJ1 are markers of nerve regeneration. To further investigate the nerve regeneration effect of the NSC-exo in cutaneous wounds, the skin sections were stained with gap43 and TUJ1 by immunofluorescence. As shown in Figure 4E and Figure S1, the areas of epithelium and deep dermis were selected in each group for photography. The results showed that the expression of gap43 in NSC-exo group and N/HA-NSC-exo group was increased. However, the expression of TUJ1 was not obvious in all groups, and some overlapped with gap43, indicating that there were few mature neurons at that time. In addition, pacinian corpuscles appeared in the deep dermis of iPSC-exo, NSC-exo, and N/HA-NSC-exo groups. Among them, the pacinian corpuscles in N/HA-NSC-exo group were obviously larger. The pacinian corpuscles are sensory nerve endings, surrounded by connective tissue to form a capsule, with a diameter of about 400–1000 μm mainly located in the deep dermis,31 the functions of which are mainly sensing mechanical sensations such as pressure, vibration and tension. The recovery of this tissue reflects the nerve recovery at the wound. These results indicate that NSC-exo could promote nerve regeneration, thus promoting wound healing. The administrations of hydrogel were likely to increase the nerve regeneration effect of NSC-exo via controlled release, and it might play an important role in wound healing.
Proteomic Analysis of NSC-Exo
To explore the potential mechanism of NSC-exo on wound healing, non-labeling proteomic analysis of the NSC-exo was performed, in which the protein of NSCs was used as the control group for comparison. As shown in Figure 5A, setting FDR <5% as the confidence interval, 1,488 significant proteins were identified in the exosomes. 46 proteins were highly expressed in the exosomes but not found in the cells, and 1,442 proteins were co-expressed in NSCs and NSC-exo with different expression levels. After normalizing the abundance value of NSCs (control) group and NSC-exo (sample) group, the highly expressed differential proteins with the abundance ratio (sample/control) >2, P<0.05 were screened. The results showed that 1471 proteins in exosomes could be quantified, and 175 proteins of them were highly expressed differential proteins (Figure 5B). Among all proteins in cells and exosomes, 753 proteins were not found or low-expressed in exosomes. A visual volcanic diagram of all proteins was shown in Figure 5C. Red dots represented highly expressed proteins in exosomes and blue dots represented low expressed proteins. In order to explore the components and functions of highly expressed differential proteins in exosomes, GO analysis was carried out on these 175 proteins by using DAVID Knowledgebase (https://david.ncifcrf.gov/). As shown in Figure 5D, the components of differential proteins were mainly extracellular secretion components, cytoplasmic membrane, extracellular region components and cytosol, which were consistent with the characteristics of exosomes. Besides, the typical protein markers with high expression identified by mass spectrometry in exosomes were listed in Table 2, including ALIX, TSG101 and four transmembrane proteins CD31. The enriched biological processes, ECM remodeling, cell adhesion, immune response, cell proliferation and cell migration were important in the process of skin wound healing, indicating that NSC-exo contained more functional proteins than NSCs.
 |
Table 2 Representative Protein Markers Identified in Exosomes by Mass Spectrometry |
As vesicles were secreted by cells, exosomes carry many kinds of functional proteins, which can be used as a natural drug repository. Therefore, the analysis of all NSC-exo proteins is helpful to understand the overall function of NSC-exo. Firstly, the GO pathway enrichments were classified according to the cell composition, biological function and molecular function of 1,488 proteins identified in exosomes by using DAVID Knowledgebase (https://david.ncifcrf.gov/). Besides various membrane components such as exosomes and cytosolic components, other cellular components such as nuclei and mitochondria, were enriched in Figure 5E. The molecular functions of differential proteins are mainly classified into protein binding, RNA binding and enzyme binding. The biological pathways involved by proteins mainly include intercellular adhesion, protein translation, redox process and so on. Pathway analysis of proteins in NSC-exo was carried out through the Panther database (http://pantherdb.org/) and the top 12 pathways were showed in Figure 5F. Among them, cytokine mediated inflammatory response pathway was related to the inflammatory phase in skin wound healing, while Wnt pathway and CCKR pathway were related to cell proliferation, migration and adhesion. Angiogenesis pathway and platelet-derived growth factor (PDGF) pathway could participate in angiogenesis and were related to the proliferation phase in wound repair.
Although there are thousands of proteins in NSC-exo, the expression abundance of different proteins varies greatly. The analysis of the top 100 relative abundant proteins (listed in Table S1) in exosomes was further carried by using the Panther database (http://pantherdb.org/), and the proteins related to skin wound repair and nerve regeneration were selected. The main abundant proteins were listed in Table 3. The results showed that gal-3bp, Agrin, hemopexin, nidogen-1, and other proteins were widely expressed in central neurons and microvascular basement membrane were also found in the top 100 proteins expressed in exosomes, which were consistent with a recent related study,19 and also played an important role in synaptic regeneration and plasticity. In addition, exosomes contain a variety of proteins related to axon regeneration, such as plexin-A2, netrin-1, and ephrin-B2. In skin wounds, axon regeneration played an important role in the repair of peripheral nerve injury and the development of neuronal regeneration. For example, Eph pathway might participate in the cascade reaction of injured cells and play an important role in axon guidance and synapse formation. Perrinello et al found that EphB signal pathway could mediate the peripheral nerve regeneration,32,33 netrin-1 and ephrin could cooperate to participate in axon guidance.34 In addition, some proteins in exosomes were involved in angiogenesis, cell migration, immune response and cell matrix remodeling, such as hemopexin, bone morphogenetic protein (BMP) and neuro-derived neurotrophic factor (NDNF). Interestingly, NDNF was a neuro-derived neurotrophic factor, which had been found that it had a unique new function in the process of vascular formation of endothelial cells, and a new target of ischemic disease.35,36 NDNF was first found in the NSC-exo in this paper, which might play an important role in wound healing. Besides, NSC-exo contained a variety of cell adhesion proteins, such as LAMB1 and syndecan-2, which may be the main proteins involved in promoting migration, while the pathways related to cell migration in NSC-exo include Wnt pathway and PI3K/Akt pathway, which remained to be verified in a molecular level. The proteins involved in angiogenesis in the proteomic analysis of NSC exosomes include S1PR1, ephrin, SFRP1 and NDNF, and the related pathways include Wnt pathway, PI3K/Akt and PDGF pathway, which still need to be further verified. Recent study shows that the existence of some proteins such as agrin and hemopexin, presented in Table 3, have been validated in iPSC-NSC-EVs by quantitative ELISAs,19 which may pave a valuable path to our validation work in the future.
 |
Table 3 The Main Proteins Among the Top 100 Relative Abundant Proteins Related to Wound Repair and Nerve Regeneration in NSC-exo |
Furthermore, our future work will investigate the therapeutic effects and related mechanisms of NSC exosomal miRNAs on wound healing. Since NSC exosomes also contain miRNAs,19,37,38 which may contribute to the therapeutic effects of exosomes. However, so far, their relevant roles on wound healing have not been explored. But we can speculate the therapeutic effects from other related studies. Based on high-throughput sequencing, Wang et al found hypoxia adipose stem cell-derived exosomes contained wound healing correlated miRNAs, which might regulate cell metabolism, differentiation and transforming growth factor-β function according to bioinformatics analysis.39 Gao et al reported that human amnion mesenchymal stem cell-derived exosomal miRNA, miR-135a, could significantly promote wound healing of rats by downregulating the expression of large tumor suppressor kinase 2.40 Another group found that miR-21 mimics, delivered by human adipose stem cell-derived exosomes, could accelerate healing effects on diabetic chronic wounds treatment by increasing angiogenesis, re-epithelialization, collagen remodeling and vessel maturation in vivo.41 In general, stem-cell derived exosomal miRNAs exert considerable therapeutic effect on wound healing.
Conclusion
In summary, we revealed the therapeutic effects of the NSC-exo on wound healing in vitro and in vivo. NSC-exo promoted the cell migration of HDF cells and tube formation of HUVECs in vitro and the wound healing in mice skin injury model in vivo. The potential mechanism of which may relate to the inflammatory regulation and the function of neural growth factors such as NDNF.
Consent for Publication
All the authors consent for publication.
Acknowledgments
We thank Professor Kaijie Chen from Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences (Guangzhou, China), for kindly providing human iPSCs. Our work was funded by National Natural Science Foundation of China (Grant no. 81971081), The Guangdong Natural Science Foundation (2018A030310097), The Innovation and Technology Fund of Guangzhou (20180310090), Shenzhen Science and Technology Program (Grant no. KQTD20190929173853397).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sun BK, Siprashvili Z, Khavari PA. Advances in skin grafting and treatment of cutaneous wounds. Science. 2014;346(6212):941–945. doi:10.1126/science.1253836
2. Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med. 2014;6(265):1. doi:10.1126/scitranslmed.3009337
3. Blakytny R, Jude E. The molecular biology of chronic wounds and delayed healing in diabetes. Diabetic Med. 2006;23(6):594–608. doi:10.1111/j.1464-5491.2006.01773.x
4. Grey JE, Enoch S, Harding KG. ABC of wound healing - Venous and arterial leg ulcers. Br Med J. 2006;332(7537):347–350. doi:10.1136/bmj.332.7537.347
5. Yucha SEV, Tamamoto KA, Kaplan DL. The importance of the neuro-immuno-cutaneous system on human skin equivalent design. Cell Prolif. 2019;52(6). doi:10.1111/cpr.12677
6. Ashrafi M, Baguneid M, Bayat A. The role of neuromediators and innervation in cutaneous wound healing. Acta Derm Venereol. 2016;96(5):587. doi:10.2340/00015555-2321
7. Kiya K, Kubo T. Neurovascular interactions in skin wound healing. Neurochem Int. 2019;125:144–150. doi:10.1016/j.neuint.2019.02.014
8. Roosterman D, Goerge T, Schneider SW, Bunnett NW, Steinhoff M. Neuronal control of skin function: the skin as a neuroimmunoendocrine organ. Physiol Rev. 2006;86(4):1309–1379. doi:10.1152/physrev.00026.2005
9. Xu Z, Han S, Gu Z, Wu J. Advances and impact of antioxidant hydrogel in chronic wound healing. Adv Healthcare Mater. 2020;9(5):1901502. doi:10.1002/adhm.201901502
10. Su DD, Tsai HI, Xu ZX, et al. Exosomal PD-L1 functions as an immunosuppressant to promote wound healing. J Extracell Vesicles. 2020;9(1):12. doi:10.1080/20013078.2019.1709262
11. Linard C, Brachet M, Strup-Perrot C, et al. Autologous bone marrow mesenchymal stem cells improve the quality and stability of vascularized flap surgery of irradiated skin in pigs. Stem Cells Transl Med. 2018;7(8):569–582. doi:10.1002/sctm.17-0267
12. Driskell RR, Lichtenberger BM, Hoste E, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504(7479):277. doi:10.1038/nature12783
13. Cusimano M, Brambilla E, Capotondo A, et al. Selective killing of spinal cord neural stem cells impairs locomotor recovery in a mouse model of spinal cord injury. J Neuroinflammation. 2018;15(1):58. doi:10.1186/s12974-018-1085-9
14. Khan M, Nickoloff E, Abramova T, et al. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ Res. 2015;117(1):52–64. doi:10.1161/circresaha.117.305990
15. Mathiyalagan P, Liang Y, Kim D, et al. Angiogenic mechanisms of human CD34(+) stem cell exosomes in the repair of ischemic hindlimb. Circ Res. 2017;120(9):1466. doi:10.1161/circresaha.116.310557
16. Webb RL, Kaiser EE, Scoville SL, et al. Human neural stem cell extracellular vesicles improve tissue and functional recovery in the murine thromboembolic stroke model. Transl Stroke Res. 2018;9(5):530–539. doi:10.1007/s12975-017-0599-2
17. Webb RL, Kaiser EE, Jurgielewicz BJ, et al. Human neural stem cell extracellular vesicles improve recovery in a porcine model of ischemic stroke. Stroke. 2018;49(5):1248. doi:10.1161/strokeaha.117.020353
18. Rong YL, Liu W, Wang JX, et al. Neural stem cell-derived small extracellular vesicles attenuate apoptosis and neuroinflammation after traumatic spinal cord injury by activating autophagy. Cell Death Dis. 2019;10:18. doi:10.1038/s41419-019-1571-8
19. Upadhya R, Madhu LN, Attaluri S, et al. Extracellular vesicles from human iPSC-derived neural stem cells: miRNA and protein signatures, and anti-inflammatory and neurogenic properties. J Extracell Vesicles. 2020;9(1):1809064. doi:10.1080/20013078.2020.1809064
20. Li WL, Sun W, Zhang Y, et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci USA. 2011;108(20):8299–8304. doi:10.1073/pnas.1014041108
21. Sim H, Seo J-H, Kim J, et al. Quantitative proteomic analysis of primitive neural stem cells from LRRK2 G2019S-associated Parkinson’s disease patient-derived iPSCs. Life-Basel. 2020;10(12). doi:10.3390/life10120331
22. Liu G-H, Qu J, Suzuki K, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491(7425):603–607. doi:10.1038/nature11557
23. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;3:22. doi:10.1002/0471143030.cb0322s30
24. Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi:10.1038/nprot.2006.468
25. Huang Da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
26. Gazdhar A, Grad I, Tamo L, et al. The secretome of induced pluripotent stem cells reduces lung fibrosis in part by hepatocyte growth factor. Stem Cell Res Ther. 2014;5(6):123. doi:10.1186/scrt513
27. Kanazawa S, Fujiwara T, Matsuzaki S, et al. bFGF regulates PI3-kinase-Rac1-JNK pathway and promotes fibroblast migration in wound healing. PLoS One. 2010;5(8):e12228. doi:10.1371/journal.pone.0012228
28. Demidova-Rice TN, Durham JT, Herman IM. Wound healing angiogenesis: innovations and challenges in acute and chronic wound healing. Adv Wound Care. 2012;1(1):17–22. doi:10.1089/wound.2011.0308
29. DeCicco-Skinner KL, Henry GH, Cataisson C, et al. Endothelial cell tube formation assay for the in vitro study of angiogenesis. J Vis Exp. 2014;(91):e51312. doi:10.3791/51312
30. Wang L, Li J, Xiong Y, et al. Ultrashort peptides and hyaluronic acid-based injectable composite hydrogels for sustained drug release and chronic diabetic wound healing. ACS Appl Mater Interfaces. 2021;13(49):58329–58339. doi:10.1021/acsami.1c16738
31. Sames K, Halata Z, Jojovic M, et al. Lectin and proteoglycan histochemistry of feline pacinian corpuscles. J Histochem Cytochem. 2001;49(1):19–28. doi:10.1177/002215540104900103
32. Parrinello S, Napoli I, Ribeiro S, et al. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell. 2010;143(1):145–155. doi:10.1016/j.cell.2010.08.039
33. Cramer KS, Cerretti DP, Siddiqui SA. EphB2 regulates axonal growth at the midline in the developing auditory brainstem. Dev Biol. 2006;295(1):76–89. doi:10.1016/j.ydbio.2006.03.010
34. Poliak S, Morales D, Croteau LP, et al. Synergistic integration of Netrin and ephrin axon guidance signals by spinal motor neurons. Elife. 2015;(4). doi:10.7554/eLife.10841
35. Kuang XL, Zhao XM, Xu HF, et al. Spatio-temporal expression of a novel neuron-derived neurotrophic factor (NDNF) in mouse brains during development. BMC Neurosci. 2010;11:137. doi:10.1186/1471-2202-11-137
36. Ohashi K, Enomoto T, Joki Y, et al. Neuron-derived neurotrophic factor functions as a novel modulator that enhances endothelial cell function and revascularization processes. J Biol Chem. 2014;289(20):14132–14144. doi:10.1074/jbc.M114.555789
37. Zhang Y, Kim MS, Jia B, et al. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature. 2017;548(7665):52–57. doi:10.1038/nature23282
38. Upadhya R, Madhu LN, Rao S, Shetty AK. Proficiency of extracellular vesicles from hiPSC-derived neural stem cells in modulating proinflammatory human microglia: role of pentraxin-3 and miRNA-21-5p. Front Mol Neurosci. 2022;15:845542. doi:10.3389/fnmol.2022.845542
39. Wang J, Wu H, Peng Y, et al. Hypoxia adipose stem cell-derived exosomes promote high-quality healing of diabetic wound involves activation of PI3K/Akt pathways. J Nanobiotechnology. 2021;19(1):202. doi:10.1186/s12951-021-00942-0
40. Gao S, Chen T, Hao Y, et al. Exosomal miR-135a derived from human amnion mesenchymal stem cells promotes cutaneous wound healing in rats and fibroblast migration by directly inhibiting LATS2 expression. Stem Cell Res Ther. 2020;11(1):56. doi:10.1186/s13287-020-1570-9
41. Lv Q, Deng J, Chen Y, et al. Engineered human adipose stem-cell-derived exosomes loaded with miR-21-5p to promote diabetic cutaneous wound healing. Mol Pharm. 2020;17(5):1723–1733. doi:10.1021/acs.molpharmaceut.0c00177
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.