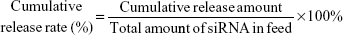Back to Journals » International Journal of Nanomedicine » Volume 10 » Issue 1
Enhanced delivery of PEAL nanoparticles with ultrasound targeted microbubble destruction mediated siRNA transfection in human MCF-7/S and MCF-7/ADR cells in vitro
Authors Teng Y, Bai M, Sun Y, Wang Q, Li F, Xing J, Du LF, Gong T, Duan Y
Received 19 January 2015
Accepted for publication 22 April 2015
Published 27 August 2015 Volume 2015:10(1) Pages 5447—5457
DOI https://doi.org/10.2147/IJN.S81172
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Lei Yang
Yanwei Teng,1,2,* Min Bai,3,* Ying Sun,2 Qi Wang,1,2 Fan Li,3 Jinfang Xing,3 Lianfang Du,3 Tao Gong,1 Yourong Duan2
1Key Laboratory of Drug Targeting and Novel Drug Delivery Systems, Ministry of Education, West China School of Pharmacy, Sichuan University, Chengdu, Sichuan, People’s Republic of China; 2State Key Laboratory of Oncogenes and Related Genes, Shanghai Cancer Institute, Renji Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, People’s Republic of China; 3Department of Ultrasound, Shanghai First People’s Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Abstract: The gene knockdown activity of small interfering RNA (siRNA) has led to their use as potential therapeutics for a variety of diseases. However, successful gene therapy requires safe and efficient delivery systems. In this study, we choose mPEG-PLGA-PLL nanoparticles (PEAL NPs) with ultrasound targeted microbubble destruction (UTMD) to efficiently deliver siRNA into cells. An emulsification-solvent evaporation method was used to prepare siRNA-loaded PEAL NPs. The NPs possessed an average size of 132.6±10.3 nm (n=5), with a uniform spherical shape, and had an encapsulation efficiency (EE) of more than 98%. As demonstrated by MTT assay, neither PEAL NPs nor siRNA-loaded PEAL NPs showed cytotoxicity even at high concentrations. The results of cellular uptake showed, with the assistance of UTMD, the siRNA-loaded PEAL NPs can be effectively internalized and can subsequently release siRNA in cells. Taken together, PEAL NPs with UTMD may be highly promising for siRNA delivery, making it possible to fully exploit the potential of siRNA-based therapeutics.
Keywords: gene delivery, mPEG-PLGA-PLL, UTMD, emulsification-solvent evaporation method, orthogonal design
Introduction
RNA interference (RNAi) is an emerging gene knockdown technology, that is more specific and efficient compared to traditional gene knockdown techniques.1–3 RNAi mediated gene silencing is induced by 21–23 nt small interfering RNAs (siRNAs), which can induce the silencing of complementary target mRNA thereby halting the corresponding protein production.4,5 However, the naked siRNA is unstable in the blood circulation due to rapid enzymatic degradation.6 Furthermore, it is difficult for siRNA to enter cells because of the negative charge of both siRNA and cell membrane.7,8 Therefore, finding an efficient siRNA-mediating system to protect siRNA against degradation and transport it to the target site is the most important hurdle for widespread use of siRNA.9 Numerous delivery vectors, both viral and nonviral, have been investigated to circumvent this problem.10 Viral vectors are highly efficient, but their clinical application is restricted by several drawbacks, including their oncogenic potential and their inflammatory and immunogenic effects, which prevent them from repeated administration.11,12 To overcome these limitations, nonviral vectors have emerged as a promising alternative for gene delivery.
Recently, cationic polymers as nonvirus gene carriers have gained significant attention because of their low cell immunogenicity and excellent structure flexibility.13 In our study, we used monomethoxy polyethylene glycol–polylactic acid/glycolic acid–poly(L-lysine) triblock copolymer (mPEG-PLGA-PLL, PEAL) with ultrasound targeted microbubble destruction (UTMD) for siRNA delivery. PEAL is a cationic polymer with a hydrophilic shell and a hydrophobic core, suitable for carrying various genes. The protonated amino group of PLL could electrostatically interact with the polyanionic siRNA and negatively charged cell membrane to increase the transportation of siRNA to the cytosol.14–16 UTMD, a safe and effective gene transfer method, can protect nanocarriers against nonspecific recognition in the systemic circulation,17–20 whereas, directional rupture of microbubbles (MB) with the help of ultrasound (US) can transiently lead to the formation of repairable pores on cell membrane, which is conducive to increase the permeability of biological macromolecules and can assist nanocarriers in penetrating cell membrane and entering into cells.21–23
In this work, the siRNA-loaded PEAL NPs were prepared by emulsification-solvent evaporation method24 and characterized in terms of morphology, size, ζ potential, and encapsulation efficiency (EE). We hypothesized that PEAL NPs with UTMD could increase the siRNA delivery efficiency. Using drug-sensitive and adriamycin-resistant human breast cancer cell strains (MCF-7/S and MCF-7/ADR) as cell models, we undertook a systematic approach to investigate the siRNA delivery efficiency and cytotoxicity of the delivery system.
Materials and methods
Materials
PEAL (30% mPEG, PLGA: LA/GA =7:3) was synthesized in our laboratory;25 Dimethyl sulfoxide (DMSO) and 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) were purchased from Sigma-Aldrich (St Louis, MO, USA); 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was purchased from Qianchen Biotechnology Inc (Shanghai, People’s Republic of China). The commercial SonoVue®, diluted with 5 mL of saline to form MB, was purchased from Bracco Diagnostics Inc (Geneva, Switzerland).
Experimental facilities of US
A Topteam 161 therapeutic US apparatus from Chattanooga Medical Supply, Inc. (Chattanooga, TN, USA) with 1 MHz in frequency, 100 Hz in pulse repetition frequency, and 25 mm2 cross-sectional area of the probe, was applied for enhancing siRNA delivery. When the insonation was performed, the US probe was placed on the bottom of the culture plate with a small amount of couplants on the surface of the probe to form the conductive pathway of US waves from transducer to cells.
Cell culture
MCF-7/S cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China) and its multidrug-resistant variant MCF-7/ADR was established and maintained in our laboratory. All cells were cultured in RPMI-1640 (Roswell Park Memorial Institute-1640) medium (Biosera, East Sussex, UK) supplemented with 10% fetal bovine serum (Biosera), and 1% penicillin and streptomycin (Biosera). MCF-7/ADR cells were grown in the RPMI-1640 complete medium with 1 μg/mL Adriamycin (ADR) so as to maintain its drug resistance. Cell cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Preparation of siRNA-loaded PEAL NPs
The siRNA-loaded PEAL NPs were prepared by emulsification-solvent evaporation method. As an example, 1 nmol of siRNA dissolved in 20 μL of RNase free water (W1) was emulsified by sonication for 50 seconds over an ice bath in 0.2 mL of dichloromethane containing 2 mg of PEAL. This W1/O (water 1/oil) primary emulsion was further emulsified in 2 mL of Pluronic 188 solution (1.0%, w/v, W2) by sonication (300 W for 2 minutes) over an ice bath to form a water-in-oil-in-water emulsion. Then the dichloromethane solvent was removed using a rotary evaporator (Taikang Biotechnology Inc, Shaanxi, People’s Republic of China) at room temperature. The resulting W1/O/W2 emulsion was stored at −20°C. The siRNA-loaded NPs were denoted as NP1.0, where NP represents nanoparticles and the subscript represents the molar weight of siRNA.
Characterization of siRNA-loaded PEAL NPs
The average particle size, size distribution, and surface potential of siRNA-loaded PEAL NPs were evaluated by a Zetasizer IV analyzer (Malvern Zetasizer Nano ZS90, Malvern Instruments, Malvern, UK). Transmission electron microscopy (TEM, JEOL, Tokyo, Japan) was used to observe the morphology of NPs. The EE of siRNA entrapped into PEAL NPs was calculated based on the concentration of the free siRNA in the filtrate obtained after ultrafiltration. The concentration of siRNA, which we labeled with cy5, was determined by a fluorescence spectrophotometer (SLM Instruments Inc, Urbana, IL, USA) at an excitation wavelength of 646 nm and an emission wavelength of 666 nm. The EE and drug loading (DL) of siRNA was calculated using the following formula:
EE (%) = (1− weight of free siRNA/siRNA) in feed ×100% |
DL (%) = Weight of siRNA in feed/weight of NPs ×100% |
In vitro release of siRNA
About 5.0 nmol of siRNAcy5 was dissolved in a PBS (phosphate buffer saline) buffer (pH 7.4) to obtain a 20 μmol/L stock solution. Then the stock solution was diluted in PBS to obtain a series of standard solutions in the range of 1–500 nmol/L.
siRNAcy5-loaded PEAL NPs (1.5 mL) were suspended in 3.0 mL of PBS in triplicate, and incubated at 37°C with gentle shaking (80 rpm). Samples were centrifuged at predetermined time intervals (2, 4, 8, 12, 24, 48, 72, 96, and 120 hours). The amount of released siRNAcy5 was determined by using the fluorescence spectrophotometer as described in the “Characterization of siRNA-loaded PEAL NPs” section. The cumulative release rate of siRNAcy5 was calculated using the following formula:
|
Cytotoxicity assay
The cytotoxicity of siRNA and PEAL was assessed by MTT assay against MCF-7/S, MCF-7/ADR, and L929 cells. Cells were seeded into 96-well plates at a density of 6×103 cells/well and cultured for 24 hours in 100 μL of RPMI-1640 complete medium. The medium was replaced with 100 μL RPMI-1640 complete medium containing different concentrations of free siRNA, blank PEAL NPs, and siRNA-loaded PEAL NPs, control group with PBS and further cultured for 24, 48, and 72 hours. After cells were incubated with MTT solution (0.5 mg/mL) for 4 hours at 37°C, with the exception of the wells as blank control, the medium was replaced with 150 μL of DMSO to dissolve the formazan product. The absorbance was measured at 570 nm using a microplate reader (Bio-Rad Laboratories Inc, Hercules, CA, USA). The cell viability was normalized to that of cells cultured in the culture medium with PBS treatment.
Optimization of delivery condition
The condition of NPs delivery mediated by UTMD was optimized through L9 (34) orthogonal design (Table 1), in which have four influencing factors (the MB: 10–30 μL, US power: 1.0–1.5 W, US exposure time: 40–60 seconds, and US duty cycle: 10%–50%) that were divided into three levels.
  | Table 1 L9 (34) orthogonal design of influencing factors |
Cellular uptake of siRNA-loaded PEAL NPs
In this study, the cells were divided into the following two groups: 1) the free siRNA group and 2) the siRNA-loaded PEAL NPs group. Each group was then divided into two subgroups: 1) no US and MB group and 2) the UTMD group.
MCF-7/S and MCF-7/ADR cells were seeded in 96-well plates at a density of 6×103 cells/well and incubated overnight. After cells reached 80% confluence, the culture medium was replaced by the desired siRNA formulations (diluted by RPMI-1640 complete medium) in the absence or presence of UTMD. siRNA labeled with cy5 was used to examine the uptake of cells. The final concentration of siRNA was 50 nmol/L. At various times of incubation, the culture medium was discarded. Then, the cells were washed with PBS three times and fixed with 4% paraformaldehyde before imaging, and the nuclei were stained with DAPI. Fluorescence inverted microscope (Huarui Chemical Instrument Inc, Guangdong, People’s Republic of China) and confocal laser scanning microscope (CLSM, Leica Inc, Allentown, NJ, USA) were used to observe the cellular uptake of siRNA.
MCF-7/S and MCF-7/ADR cells were seeded in six-well plates at a density of 1×105 cells/well. After 24 hours, the culture medium was replaced by the desired siRNA formulations. At 4 hours of incubation, the culture medium was discarded. Then, the cells were washed with PBS three times and harvested by trypsinization. The samples were analyzed by flow cytometry (BD Bioscience, San Jose, CA, USA) to determine the transfection efficiency of siRNA quantitatively.
siRNA-mediated silencing efficiency
The overexpression of the breast cancer resistance protein (ABCG2) confers multidrug resistance in cancers. In this study, we chose ABCG2-siRNA as the model siRNA (sense: 5′-CUGGAGAUGUUCUGAUAAA dTdT-3′, antisense: 3′-dTdT GACCUCUACAAGACUAUUU-5′). MCF-7/S and MCF-7/ADR cells were seeded onto six-well plates at a density of 1×105 cells/well, incubated for 24 hours, and subsequently treated with different formulations containing 100 nM ABCG2-siRNA or negative control-siRNA (NC-siRNA) in the absence or presence of UTMD. Untreated cells were used as a negative control. After incubation for 48 hours, total protein or mRNA was extracted from the cells for Western blotting or reverse transcription PCR (RT-PCR) analysis, respectively, to assess ABCG2 protein and gene expression levels.
Statistical analysis
All data analyses were performed in triplicate and the data were shown as the mean ± standard deviation (SD). One-way analysis of variance (ANOVA) was used to analyze results. GraphPad Prism (GraphPad Software Inc, San Diego, CA, USA) was used to obtain graphs and statistics. Significant values were designated as follows: *P<0.05, **P<0.01, and ***P<0.001.
Results and discussion
Preparation and characterization of siRNA-loaded PEAL NPs
In this study, we selected PEAL as the nanocarrier. The emulsification-solvent evaporation method is shown in Figure 1. The cationic polymer can form NPs, and the siRNA is entrapped within the NP via ionic interactions and not by simple adsorption onto the surface,13 therefore, siRNA could be immobilized in the formed NPs at high EE. Furthermore, the outer layer of siRNA-loaded PEAL NPs was covered with mPEG, which could improve the stability of the NPs due to its hydrophilicity and excellent biocompatibility.26,27
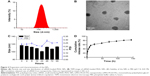
As summarized in Table 2, the ζ potentials of siRNA-loaded PEAL NPs were found to be negatively correlated with the amount of siRNA in feed. Recent data show that NPs carrying a very slight positive charge can penetrate throughout large tumors following systemic administration.28 Additionally, based on the EE and DL, NP1.0 was used for subsequent experiments. The average hydrodynamic diameter (Figure 2A) and the TEM image (Figure 2B) showed that NP1.0 possessed a spherical nanostructure with no aggregation and dispersed well. To investigate the stability of the NPs, the NPs were incubated in PBS (0.01 M, pH =7.4) at 37°C and the average size of NPs was monitored subsequently for 96 hours. As shown in Figure 2C, the size of NPs remained constant during the whole incubation period, especially within 12 hours incubation. The results demonstrated that the hydrated shell formed by mPEG could enhance the hydrophilicity of the NPs and keep them stable at least for 96 hours.
The siRNA concentration and its absorbance value had a linear relationship in the 4–500 nM concentration range of siRNA. The standard curve equation of siRNAcy5 was as follows: A =0.5739C−0.0212, r2 =0.9997, where A represented the absorbance of siRNAcy5 at an excitation wavelength of 646 nm and an emission wavelength of 666 nm, and C represented the concentration of siRNAcy5. The in vitro release experiments were carried out at 37°C with a constant shaking rate of 80 rpm. As shown in Figure 2D, the NPs exhibited a burst release of ~28% at 12 hours, which may be attributable to the siRNA being adsorbed on the surface of NPs.29 Then, with the degradation of PEAL, the siRNA entrapped in the NPs was gradually released and exhibited sustained release patterns over an extended period. We all know that siRNA is greatly constricted by its instability and quick degradation in the body. These results indicated that PEAL NPs had good sustained release properties and were expected to evade the weakness of siRNA and prolong its circulation time in vivo.
Cytotoxicity
In order to evaluate the in vitro toxicity of siRNA and PEAL, cytotoxicity studies were conducted by using various doses of free siRNA, blank PEAL NPs, and siRNA-loaded PEAL NPs against MCF-7/S, MCF-7/ADR, and L929 cells for 24, 48, and 72 hours. As shown in Figure 3, all groups maintained over 80% cell viability for dosage up to 120 nmol/L within 72 hours, suggesting their good biocompatibility. In addition, a comprehensive evaluation has already been done before to evaluate the safety of PEAL.30 Therefore, the PEAL NPs could be safely used for future studies and in vivo studies.
Orthogonal design and optimized composition
The optimization results of orthogonal test are shown in Table 3. The results demonstrated that the optimized experimental conditions of cellular uptake and mean fluorescence intensity (MFI) were identical. The optimization parameters were as follows: 20% MB, 1.0 W US power, 40 seconds US exposure time, and 20% duty cycle.
Cellular uptake of siRNA-loadedPEAL NPs
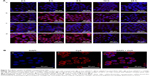
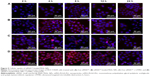
Internalization of different siRNAs was evaluated by fluorescence inverted microscope and flow cytometry. To understand how the incubation time affected cell internalization of siRNA, we incubated siRNAcy5 and siRNAcy5-loaded NPs with MCF-7/S and MCF-7/ADR cells for different periods of time in the absence or presence of UTMD and subsequently observed the intracellular fluorescence intensity by a microscope. As shown in Figures 4 and 5, incubation of siRNAcy5 alone exhibited very minimal cell fluorescence, while significantly enhanced fluorescence intensity was observed in cells when siRNA was entrapped into PEAL NPs, and the fluorescence intensity further enhanced after the formulations were combined with UTMD, indicating that combining UTMD with NPs could facilitate the entry of siRNA into cells effectively, which was mainly because the sonoporation induced by UTMD could promote siRNA penetration through cell membrane. Inverted fluorescence microscope was used to observe the intracellular distribution of siRNA more precisely. The results (Figure 4B) demonstrated that most of the siRNA was distributed in the cytoplasm, which is consistent with other groups’ observation.31,32
To further investigate the uptake effects of MCF-7/S and MCF-7/ADR cells toward siRNA, flow cytometry was used to determine the intracellular fluorescence intensity of siRNAcy5 at 4 hours after transfection. Flow cytometric analysis (Figure 6) showed that more than 95% of cells treated with siRNAcy5-loaded PEAL NPs were positive for fluorescence as compared with untreated cells. The MFI of MCF-7/S cells treated with free siRNA and siRNA-loaded NPs were ~50 arbitray units (AU) and ~554 AU, respectively, and the corresponding UTMD groups were ~102 AU and ~727 AU, respectively; MCF-7/ADR cells treated with free siRNA and siRNA-loaded NPs were ~54 AU and ~471 AU, respectively, and the corresponding UTMD groups were ~83 and ~652, respectively. The MFI of siRNA-loaded NPs was significantly higher than that of free siRNA (P<0.001), and the MFI of US groups were all higher than nonUS groups (P<0.05), which was in agreement with qualitative analyses of fluorescence microscopy. Naked siRNA is unstable and easily degraded, which greatly limits its application. Therefore, combining PEAL NPs with UTMD could protect siRNA from degradation and improve its cell uptake efficiency, making it possible to fully exploit the potential of siRNA-based therapeutics.
siRNA-mediated silencing efficiency
MCF-7/S and MCF-7/ADR cells were incubated with different siRNA formulations, and the level of ABCG2 mRNA was analyzed after 48 hours through RT-PCR. As shown in Figure 7A, NC-siRNA did not significantly knockdown ABCG2 mRNA expression compared with the control, further confirming the sequence specificity of gene silencing through siRNA. Compared with naked siRNA, siRNA-loaded PEAL NPs significantly downregulated ABCG2 mRNA expression, whereas, in the presence of UTMD, siRNA/PEAL NPs exhibited an enhanced knockdown effect.
Western blot analysis was also performed to detect ABCG2 expression at the protein level. As shown in Figure 7B, the delivery of ABCG2-siRNA through PEAL NPs + UTMD exhibited the best suppression effect on ABCG2 protein expression, consistent with the results of RT-PCR.
Conclusion
In this study, PEAL NPs were used as gene vectors because of their various advantages. MTT assay demonstrated it had good cytocompatibility even at high concentrations, suitable for carrying various genes. The PEAL NPs, along with UTMD, could protect siRNA from degradation and mediate effective siRNA delivery. In conclusion, PEAL NPs with UTMD may be highly promising for siRNA delivery, making it possible to fully exploit the potential of siRNA-based therapeutics.
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (numbers 81201097, 81272568, 81472841, and 81301973) and the Nature Science Foundation of Shanghai (number 12ZR1424800).
Disclosure
The authors report no conflicts of interest in this work.
References
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. | ||
Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95(26):15502–15507. | ||
Takahashi Y, Yamaoka K, Nishikawa M, Takakura Y. Quantitative and temporal analysis of gene silencing in tumor cells induced by small interfering RNA or short hairpin RNA expressed from plasmid vectors. J Pharm Sci. 2009;98(1):74–80. | ||
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. | ||
McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3(10):737–747. | ||
Rettig GR, Behlke MA. Progress toward in vivo use of siRNAs-II. Mol Ther. 2012;20(3):483–512. | ||
Wullner U, Neef I, Tur MK, Barth S. Targeted delivery of short interfering RNAs – strategies for in vivo delivery. Recent Pat Anticancer Drug Discov. 2009;4(1):1–8. | ||
Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010;277(23):4814–4827. | ||
Meng Q, Yin Q, Li Y. Nanocarriers for siRNA delivery to overcome cancer multidrug resistance. Chin Sci Bull. 2013;58:4021–4030. | ||
Li SD, Huang L. Non-viral is superior to viral gene delivery. J Control Release. 2007;123(3):181–183. | ||
Kim WJ, Kim SW. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm Res. 2009;26(3):657–666. | ||
Schagen FH, Ossevoort M, Toes RE, Hoeben RC. Immune responses against adenoviral vectors and their transgene products: a review of strategies for evasion. Crit Rev Oncol Hematol. 2004;50(1):51–70. | ||
Zhang S, Zhao B, Jiang H, Wang B, Ma B. Cationic lipids and polymers mediated vectors for delivery of siRNA. J Control Release. 2007;123(1):1–10. | ||
Akhtar S, Benter I. Toxicogenomics of non-viral drug delivery systems for RNAi: potential impact on siRNA-mediated gene silencing activity and specificity. Adv Drug Deliv Rev. 2007;59(2–3):164–182. | ||
Ebbesen M, Jensen TG. Nanomedicine: techniques, potentials, and ethical implications. J Biomed Biotechnol. 2006;2006(5):1–11. | ||
Kawakami S, Hashida M. Targeted delivery systems of small interfering RNA by systemic administration. Drug Metab Pharmacokinet. 2007;22(3):142–151. | ||
Chappell JC, Song J, Burke CW, Klibanov AL, Price RJ. Targeted delivery of nanoparticles bearing fibroblast growth factor-2 by ultrasonic microbubble destruction for therapeutic arteriogenesis. Small. 2008;4(10):1769–1777. | ||
Vancraeynest D, Havaux X, Pouleur AC, et al. Myocardial delivery of colloid nanoparticles using ultrasound-targeted microbubble destruction. Eur Heart J. 2006;27(2):237–245. | ||
Li HL, Zheng XZ, Wang HP, Li F, Wu Y, Du LF. Ultrasound-targeted microbubble destruction enhances AAV-mediated gene transfection in human RPE cells in vitro and rat retina in vivo. Gene Ther. 2009;16(9):1146–1153. | ||
Carson AR, McTiernan CF, Lavery L, et al. Gene therapy of carcinoma using ultrasound-targeted microbubble destruction. Ultrasound Med Biol. 2011;37(3):393–402. | ||
Liang HD, Tang J, Halliwell M. Sonoporation, drug delivery, and gene therapy. Proc Inst Mech Eng H. 2010;224(2):343–361. | ||
Zhou Y, Kumon RE, Cui J, Deng CX. The size of sonoporation pores on the cell membrane. Ultrasound Med Biol. 2009;35(10):1756–1760. | ||
Wu J, Nyborg WL. Ultrasound, cavitation bubbles and their interaction with cells. Adv Drug Deliv Rev. 2008;60(10):1103–1116. | ||
Liu P, Qin L, Wang Q, et al. cRGD-functionalized mPEG-PLGA-PLL nanoparticles for imaging and therapy of breast cancer. Biomaterials. 2012;33(28):6739–6747. | ||
Liu P, Yu H, Sun Y, et al. A mPEG-PLGA-b-PLL copolymer carrier for adriamycin and siRNA delivery. Biomaterials. 2012;33(17):4403–4412. | ||
Deshmukh M, Kutscher HL, Gao D, et al. Biodistribution and renal clearance of biocompatible lung targeted poly(ethylene glycol) (PEG) nanogel aggregates. J Control Release. 2012;164(1):65–73. | ||
Wang YY, Lu LX, Shi JC, Wang HF, Xiao ZD, Huang NP. Introducing RGD peptides on PHBV films through PEG-containing cross-linkers to improve the biocompatibility. Biomacromolecules. 2011;12(3):551–559. | ||
Davis ME, Chen ZG, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7(9):771–782. | ||
Shi Q, Liu P, Sun Y, et al. siRNA delivery mediated by copolymer nanoparticles, phospholipid stabilized sulphur hexafluoride microbubbles and ultrasound. J Biomed Nanotechnol. 2014;10(3):436–444. | ||
He Z, Wang Q, Sun Y, et al. The biocompatibility evaluation of mPEG-PLGA-PLL copolymer and different LA/GA ratio effects for biocompatibility. J Biomater Sci Polym Ed. 2014;25(9):943–964. | ||
Savic R, Luo L, Eisenberg A, Maysinger D. Micellar nanocontainers distribute to defined cytoplasmic organelles. Science. 2003;300(5619):615–618. | ||
Sun TM, Du JZ, Yan LF, Mao HQ, Wang J. Self-assembled biodegradable micellar nanoparticles of amphiphilic and cationic block copolymer for siRNA delivery. Biomaterials. 2008;29(32):4348–4355. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.