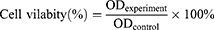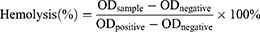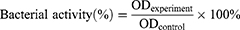Back to Journals » International Journal of Nanomedicine » Volume 16
Enhanced Antibacterial and Anti-Biofilm Activities of Antimicrobial Peptides Modified Silver Nanoparticles
Authors Xu J, Li Y, Wang H, Zhu M, Feng W, Liang G
Received 20 April 2021
Accepted for publication 16 June 2021
Published 16 July 2021 Volume 2021:16 Pages 4831—4846
DOI https://doi.org/10.2147/IJN.S315839
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 6
Editor who approved publication: Professor Israel (Rudi) Rubinstein
Juanjuan Xu,1,* Yuanpei Li,1,* Haojie Wang,1 Mengxi Zhu,1 Wenpo Feng,2 Gaofeng Liang1
1School of Basic Medical Sciences, Henan University of Science and Technology, Luoyang, 471023, People’s Republic of China; 2Medical College, Pingdingshan University, Pingdingshan, 467000, Henan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Gaofeng Liang
School of Basic Medical Sciences, Henan University of Science and Technology, Luoyang, 471023, People’s Republic of China
Email [email protected]
Wenpo Feng
Medical College, Pingdingshan University, Pingdingshan, 467000, Henan, People’s Republic of China
Email [email protected]
Background: The biofilms could protect bacteria from antibiotics and promote the production of drug-resistant strains, making the bacteria more difficult to be eradicated. Thus, we developed an AMP@PDA@AgNPs nanocomposite, which is formed by modifying silver nanoparticles (AgNPs) with antimicrobial peptides (AMP) modified nanocomposite to destroy biofilm in this study.
Methods: The AMP@PDA@AgNPs nanocomposite was prepared with polymerization method and characterized by using ultraviolet-visible (UV-vis) spectroscopy, dynamic light scattering (DLS), Fourier transform-infrared spectroscopy (FT-IR), and transmission electron microscope (TEM). The antibacterial effects of the nanocomposite were investigated by using agar diffusion method and minimum inhibitory concentration (MIC) test. The quantitative analysis of the biofilm formation by the nanocomposite was conducted using crystal violet staining and confocal laser scanning microscope (CLSM).
Results: The DLS and TEM analysis showed it was a spherical nanocomposite with 200 nm size and well dispersed . The results of UV-vis and FT-IR confirmed the presence of AMP and AgNPs. The nanocomposite had an excellent biocompatibility at 100 μg/mL. And the AMP@PDA@AgNPs nanocomposite showed superior antimicrobial activity against both Gram-negative (E. coli, P. aeruginosa) and Gram-positive (S. aureus) bacteria than AgNPs or AMP. Importantly, the mRNA expression of biofilm-related genes were decreased under the action of the nanocomposites.
Conclusion: An AMP@PDA@AgNPs nanocomposite with good biocompatibility was successfully prepared. The nanocomposite could destruct bacterial biofilms by inhibiting the expression of biofilm-related genes. The synergistic strategy of AMPs and AgNPs could provide a new perspective for the treatment of bacterial infection.
Keywords: silver nanoparticle, antimicrobial peptides, biofilm, bacteria
Introduction
Bacterial infection is one of the hardest conundrum to deal with in the world, which poses serious threat to human health. More than 90% of bacteria grow to form biofilms naturally,1 which is a microbial community that irreversibly adheres to the surface of material or tissue by self-secreted extracellular polymeric substances (EPS).2 EPS include exopolysaccharides, proteins, and extracellular DNA (eDNA),3–5 which serves as a natural barrier against the penetration of antibiotics and cellular attack by host innate immune cells.6,7 Biofilm formation involves a series of complex processes, including surface attachment of planktonic bacteria, proliferation of adherent cells and production of EPS, formation of small colonies and mature biofilm, and diffusion of mature biofilms.8,9 Bacteria living in biofilms can resist harsh living conditions and aggravate chronic infections compared with planktonic bacteria.10 It is well known that antibiotic can effectively suppress the growth of bacteria, but it is challenging to eradicate all the bacteria living under the biofilms.11 The resistance to traditional antibiotics makes biofilm-related infections more difficult to be treated compared to planktonic bacteria.12 Therefore, there is an urgent need to design and develop novel bactericides to effectively treat biofilm infections caused by bacteria.
Nowadays, nanomaterials have been widely used in the fields of biomedicine, cosmetics industry and environmental management because of their unique physical and chemical properties, strong bactericidal effect.13,14 These characteristics of nanomaterials different from conventional antimicrobial agents provide a new direction for preventing and even eradicating biofilm formation.15 For example, most metal nanomaterials can release metal ions to inactivate bacteria, such as silver nanoparticle (AgNPs),16 zinc oxide nanoparticles,17 iron oxide nanoparticles.18 It is essential to develop more effective antibacterial drugs to treat bacterial infections caused by both Gram-negative and Gram-positive pathogens. Among these nanomaterials, AgNPs have drawn most attention due to its broad-spectrum antibacterial property and low bacterial resistance.19,20 AgNPs exert their antimicrobial activity by disrupting bacterial cell membranes and continuous release of Ag+ ions. Ag+ ions can cause oxidative stress and damage DNA in bacterial cells.21 Nevertheless, the excessive leakage of metal ions can result in the toxicity to the organism22 and cause damage to local tissue side effects at high dosages. On the other hand, single silver nanoparticles tend to aggregate due to their small particle size and high surface potential.23 AgNPs are easy to be oxidized in practical applications, resulting in loss of antimicrobial activity. To settle this problem, different methods have been used to modify AgNPs.24–26 For instance, Ramyaa et al reported that dexamethasone-silver nanoparticles embedded in dendritic collagen matrix nanoparticles reduced biofilm formation in Staphylococcus aureus and Klebsiella pneumonia.26
With the rapid development of nanotechnology, a lot of novel antimicrobial agents and methods have been put forward and studied, such as antibacterial peptides (AMP),27,28 quaternary ammonium compounds,29 quantum dots,30 photodynamic therapy31 and photothermal therapy.32 Antimicrobial peptides are a diverse group of naturally occurring molecules, which can be produced by various kinds of living organisms including bacteria and animals.33 They are increasingly being considered as useful alternatives to conventional antibiotics because of their potent membrane-targeting.34 Recent studies show the thermoplastic polyurethane (TPU) surfaces modified with the peptide polymer exhibited broad-spectrum antibacterial property as well as excellent biocompatibility.35 However, the development and application of AMPs still face some challenges, such as its toxicity to eukaryotic cells and sensitivity to enzyme.36,37
Polydopamine (PDA) is an oxidative polymerization product of dopamine,38 which exhibits excellent adhesion properties and cytocompatibility.39 In an interesting study, PDA, as a simple and effective chemical surface modifier, could be developed and applied in targeted delivery systems.40 Therefore, we modified AgNPs with AMP using PDA as green reducing agent and adhesive to enhance antibacterial ability. This nanocomposite has the potential to improve the stability of single silver ions, taking full advantage of the penetration properties of antimicrobial peptides. The synthesis route of AMP@PDA@AgNPs nanocomposite by combing AgNPs and AMP is illustrated in Figure 1.
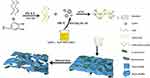 |
Figure 1 Scheme of the synthesis antibacterial AMP@PDA@AgNPs. |
Experimental Section
Materials
Sodium citrate, silver nitrate (AgNO3, 99.99%), dopamine hydrochloride (DA), crystal violet, fluorescein isothiocyanate (FITC), dimethyl sulfoxide (DMSO), and ethidium bromide were purchased from Aladdin Reagent Co., Ltd. (Shanghai, China). Tryptone, yeast, sodium chloride, ethanol, chloroform, and isopropanol were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Gram-positive bacteria Staphylococcus aureus ATCC 25923 (S. aureus), Gram-negative bacteria Escherichia coli ATCC 25922 (E. coli) and Pseudomonas aeruginosa (P. aeruginosa) were obtained from Henan Provincial Center for Disease Control and Prevention.
Synthesis of AMP@PDA@AgNPs
In a typical preparation,41 0.5 mL AMP (100 mg/mL) was added into 20 mL of Tris-HCl solution (10 mM, pH8.5) containing 1 mg of dopamine hydrochloride. The above solution was ultrasonic treated for 40 min, then stirred continuously at 25°C for 1 h. After centrifugation at 12,000 rpm for 10 min, the composites were collected and dissolved in deionized water. In this process, the AMP was modified with PDA to form AMP@PDA.
Spherical AgNPs were synthesized stably using sodium citrate as reducing agent and stabilizer.42 Briefly, 50 mL of AgNO3 solution (1 mM) was heated to boiling; then, 5 mL sodium citrate solution (1% W/V) was added drop-wisely. The solution was heated by stirring continuously at boiling point until the color changed to yellowish. After the reaction was completed, the product was named as AgNPs.
AMP@PDA@AgNPs were synthesized following a previous method.43 Briefly, 6 mL AMP@PDA (5 mg/mL) suspension was mixed with 20 mL deionized water; then, 0.5 mL ammonia solution (28 wt%) were added into the above solution with stirring for 20 min. After that, 1 mL AgNPs solution (5 mg/mL) was added to the above mixture and stirred continuously for 1 h to obtain AMP@PDA@AgNPs.
Characterization of AMP@PDA@AgNPs
The size distribution of the materials was measured using a Zetasizer Nano ZS instrument (Zetasizer, Malvern, UK). The Ultraviolet-visible (UV-vis) absorption spectra was obtained using a Shimadzu 3100 UV-2505 spectrometer (Shimadzu, Japan). The UV-vis absorption spectra of liquid nanocomposite was directly measured in a quartz glass colorimetric dish. Fourier transform-infrared spectroscopy (FT-IR) spectra of AMP, AMP@PDA, AMP@PDA@AgNPs was recorded on a Nicolet 200 type Fourier transform infrared spectrometer (Thermo Nicolet, Wilmington, USA). The liquid nanocomposite was precooled at low temperatures and then freeze-dried to make powder samples. The sample was detected by Fourier transform infrared spectrometer after KBr compression method. The result of FTIR spectrum was analysed by reading a large number of references. The morphology and size of nanocomposite were measured by transmission electron microscope (TEM, HITACHI, Japan). 10μL the liquid nanocomposite was dropped onto the copper mesh, which was naturally dried and observed under transmission electron microscope.
Safety Capability Evaluation
The human embryonic kidney 293T (HEK293T) cell line was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cytotoxicity of nanocomposite was assessed by methyl thiazolyl tetrazolium (MTT) viability assay. In brief, the cells were cultured with DMEM (Dulbecco’s modification of eagle’s medium) containing 10% fetal bovine serum (FBS) in a 96-well plates (NEST, Wuxi, China) at 37°C in a 5% CO2 humidified incubator (Thermo Fisher Scientific, Wilmington, USA). After 12 h incubation, the supernatant was removed and 200 µL DMEM with different concentrations of AgNPs, AMP, AMP@PDA@AgNPs (0, 25, 50, 100, 200, 400 µg/mL) were added to the 96-well plates. After 24 h incubation at 37°C, 15 μL of MTT (5 mg/mL) was added to each well and the supernatant was taken out after culturing for 4 h. Thereafter, 150 μL of Dimethyl sulfoxide (DMSO) was added to each well and shaken until the crystal was completely dissolved. Finally, the cell viability was determined by measuring the optical density (OD) of the samples at 490 nm on a microplate spectrophotometer (PerkinElmer, Waltham, MA, USA). The cell viability was calculated using equation (1),
For hemolysis test, the fresh blood was taken from rabbit, which was washed completely with normal saline by centrifuging at 1500 rpm for 5 times until the washing liquid became colorless. The 2% (V/V) red blood cell suspension was mixed with different concentrations AMP@PDA@AgNPs (25, 50, 100, 200, 400 µg/mL), and normal saline, pure water was designed as the negative and positive control, respectively. The mixed solution was incubated in a water bath at 37°C for 2 h, then centrifuged at 1500 rpm for 5 times. And the OD of supernatant was detected by the microplate spectrophotometer at 540 nm. The hemolysis ratio was calculated using equation (2),
In vitro Antibacterial Activity
The bacteria (E. coli, P. aeruginosa and S. aureus) preserved in glycerol were inoculated in Luria-Bertani (LB) agar plate for activation. Then, single colony was selected and cultured at 37°C in fresh LB medium. The antimicrobial efficacy of the composite against both Gram-negative (E. coli and P. aeruginosa) and Gram-positive (S. aureus) bacteria were determined by the agar diffusion test.44 The sterile circular filter papers with 6 mm diameter were fully soaked in the solution of PBS, AgNPs, AMP, AMP@PDA, AMP@PDA@AgNPs for 1 h, respectively. The concentration of AgNPs (AMP, AMP@PDA, and AMP@PDA@AgNPs) was set as 100 μg/mL. The bacterial suspension was spread on the agar plates, and the sterile circular filter papers of different treatment were placed on the agar plate. The agar plates were incubated upside down at 37°C in an incubator (Yiheng Scientific, Shanghai, China) for 12 h. After 12 h incubation, the diameters of the inhibition zone were measured to compare the bactericidal activity. Each sample was carried out in triplicates.
The antibacterial efficacy of the AMP@PDA@AgNPs nanocomposite was analyzed by detecting the bacterial activity.45 In brief, 150 μL of 1×108 CFU/mL bacterial suspension with the same volume of PBS, or 100 µg/mL of AgNPs, AMP, AMP@PDA and AMP@PDA@AgNPs were added into a 96-well plates, respectively. After incubation for 12 h, the OD value was measured at 600 nm to monitor the bacteria activity. Bacterial activity was calculated as equation (3),
The minimum inhibitory concentration (MIC) of AMP@PDA@AgNPs was determined as follows. The bacterial suspensions with the different concentrations of nanocomposite were incubated at 37°C for 24 h with shaking condition. After incubation for 3 h, 6 h, 9 h, 12 h and 24 h, the optical density of the bacterial solution at 600 nm was used to monitor the growth curve of bacteria.
Biofilms Formation and Detection
The assay for biofilms formation and detection was adapted from the previous report.46 The S. aureus suspension of different quantity (1×108, 5×107, 1×107, 5×106 CFU/mL) with 1% glucose was added into 96-well plates and incubated for 48 h to assess the biofilm formation. After incubation for 48 h at 37°C under aerobic conditions, free-floating bacteria and culture medium were removed. The biofilms were formed on the bottom of the 96-well plates, which were carefully washed twice with sterilized PBS. The biofilms were detected by semi-quantitative analysis of crystal violet staining. The S. aureus biofilms were fixed with 100 µL paraformaldehyde solution for 20 min. Afterwards, the supernatant was removed and the remaining biofilms were stained by crystal violet solution (0.1%, 20 µL) for 10 min. Then, the biofilms at the bottom of the 96-well plates were washed with deionized water for three times, and the results of crystal violet staining was obtained. In addition, crystal violet was dissolved in 100 µL ethanol for semi-quantitative analysis of biofilms, the OD values of samples were detected at 570 nm on a microplate spectrophotometer. Assays for E. coli and P. aeruginosa were conducted in the same manner as for S. aureus.
Destruction of Biofilms
The biofilms were observed by semi-quantitative analysis of crystal violet staining and the confocal laser scanning microscopy (CLSM, Carl Zeiss, Jena, Germany). The crystal violet staining procedure was similar to the biofilm formation and detection. In brief, after the biofilms were formed at the bottom of the 96-well plates, 150 μL PBS, AgNPs, AMP, AMP@PDA, AMP@PDA@AgNPs (100 μg/mL) was added into each well to co-culture with the biofilms for 12 h to detect the S. aureus biofilm biomass. Assays for E. coli and P. aeruginosa were conducted in the same manner as for S. aureus.
The bacterial biofilms were observed by CLSM, and the biofilms treated with different materials were stained with fluorescein isothiocyanate (FITC) solution. After staining for 20 min, the biofilms were washed with saline and imaged by CLSM. The 3D images of the bacteria biofilms were obtained and analyzed by using the ZEN software (Carl Zeiss, Jena, Germany).
Expression of the Genes Related to the Biofilm Formation
S. aureus, E. coli and P. aeruginosa were cultivated in six-well plate with different concentrations of AMP@PDA@AgNPs (0 µg/mL, 50 µg/mL, 100 µg/mL) at 37°C for 12 h. After forming the mature biofilms, the total RNA was extracted using TriZol Reagent (TaKaRa, Dalian, China). Next, 1 µg of the total RNA was reverse transcribed using a reverse HiFiScript cDNA synthesis kit (Cowin Bio, Beijing, China). The cDNAs were performed by 2×Taq Master Mix (Cowin Bio, Beijing, China) following the manufacturer’s instructions. The PCR assay was performed with 30 cycles at 95°C for 40 s, 54°C for 30 s, and 72°C for 30 s. The sequences of the primers used in the experiment are listed in Table 1. The genes investigated included fim H,47 a gene related with biofilm formation in E. coli, rh II and las I genes related with biofilm formation in P. aeruginosa.48 16S rRNA was used as control. The PCR products were detected by 1% agar gel electrophoresis with ethidium bromide to stain, and the bands were displayed under the ultraviolet-visible light. Subsequently, the intensity of image bands was quantitative analyzed by using Image J software.
 |
Table 1 Primer Sequences Used in PCR Amplification |
Statistical Analysis
Each experiment was repeated at least 3 times. Data was expressed as the mean ± standard deviation and analyzed using one-way analysis of variance (ANOVA). Differences between different groups at *p<0.05, **p<0.01, ***p<0.001, were considered as statistically significant difference.
Results and Discussion
Preparation and Characterization of AMP@PDA@AgNPs
The AMP@PDA@AgNPs nanocomposite was synthesized by the combination of AMPs and AgNPs (Figure 1). It was characterized by FT-IR, UV-vis, TEM and DLS. The results of DLS showed that the size of AMP, AMP@PDA, AMP@PDA@AgNPs were 79 nm, 126 nm, 150 nm, respectively (Figure 2A). These results suggested that the gradual size increase with the conjugation of AMP and AgNPs. The nanomaterials were characterized using TEM analysis (Figure 2D). TEM analysis shows that AMP@PDA was spherical or elliptical with average particle size of 35~50 nm, indicating dispersivity and stability (Figure 2D(1)). The size of AgNPs was 90~110 nm, (Figure 2D(2)). And the AMP@PDA@AgNPs was spherical and well dispersed, with a size of about 200 nm (Figure 2D(3)). The inconsistency of size between TEM and DLS measurements may be caused by the properties of nanomaterials in aqueous solutions and solids. The hydrophilic or hydrophobic properties of nanomaterials in aqueous solution are related to the size of nanomaterials measured by DLS.49 The UV-Vis absorption spectrum are shown in Figure 2B, where the AMP had an absorption peak near 230 nm due to the group of -NH3+.50 The AMP@PDA had a broad absorption peak from 200 nm to 300 nm since rich phenolic groups on the surface of PDA, which indicated the combination of AMP and PDA.44 As for AMP@PDA@AgNPs, the apparent absorption peak at 412 nm originated from the conjugation of AgNPs.51 The study by Enas et al24 showed an absorption maximum at 429 under the cupressus macrocarpa extract biosynthesis of AgNPs. Slight variation in absorption peak may be attributed to difference in the amount of the reducing chemicals present in the extracts. The above results showed that the AMP and AgNPs successfully combined to form AMP@PDA@AgNPs nanocomposite.
To further demonstrate the successful conjugation of AMP and AgNPs, the FT-IR was used to characterize different groups. As shown in Figure 2C, the characteristic peaks at near 3200 cm−1 confirmed the presence of intermolecular bonded -OH and -NH groups which signified the presence of PDA.45 In addition, the peak at 1000 cm−1 is likely due to the presence of the C-N group of PDA.52 The above results showed that the AMP@PDA and AMP@PDA@AgNPs contained the structure of PDA. The obvious absorption peak at 3100 cm−1 can be assigned to the unsaturated C-H stretching vibration of AMP. The sharp band at 1529 cm−1could possible due to N-O asymmetric stretching indicates the active involvement of nitro compounds.53 Furthermore, the presence of C=C stretch at around 1615 cm−1 confirms the presence of broad range of alkene group in the synthesized nanoparticles.44 The stretching vibration of C=C or the asymmetric stretching of N-O, which may be originated from AMP. The above results proved that AMP@PDA@AgNPs was successfully prepared.
Biological Safety Evaluation
It is vital to possess remarkable antibacterial ability while exhibiting low toxicity for practical application.54 As is known that the nanoparticles at high concentration could result in damage to organism,55 therefore the hemolytic activity and the cytotoxicity of the AMP@PDA@AgNPs nanocomposite were detected to assess biosafety. The viability of HEK293T cells was more than 95% even at the concentration of 400 µg/mL nanocomposite, implying the good biocompatibility of the nanocomposite (Figure 3A). Meanwhile, the cell viability was more than 90% after incubation with various concentrations of AMP for 24 h. However, the cell viability reduced to 8.48%, 9.53% at 200 μg/mL and 400 μg/mL AgNPs, respectively (Figure 3A). This study revealed that low AgNPs concentrations showed low toxicity in the HEK293T cell. Compared with AMP@PDA@AgNPs nanocomposite, AgNPs showed significant difference at 200 μg/mL and 400 μg/mL (**p<0.01). This result indicated that the combination of AgNPs and AMP reduced the cytotoxicity. To better evaluate the potential applications of the prepared nanocomposite, the hemolysis experiments were performed.
The hemolysis rate was only 3.59% at 100 μg/mL nanocomposite (Figure 3B), which increased to 34.46% and 56.87% at 200 μg/mL and 400 μg/mL nanocomposite, respectively (Figure 3B). These results showed that the nanocomposite had an excellent biocompatibility at 100 μg/mL. Hence, 100 μg/mL AMP@PDA@AgNPs nanocomposite was selected as a standard concentration model in this study to investigate the antibacterial efficacy.
Antimicrobial Activity
To verify the in vitro antibacterial ability of this AMP@PDA@AgNPs nanocomposite, the antibacterial effect of nanocomposite was tested against different kinds of bacteria (E. coli, P. aeruginosa and S. aureus) by the agar diffusion test. As shown in Figure 4A, none of the tested bacteria were suppressed in the PBS control group. As expected, the inhibition zone was obviously visible in all bacteria under the action of AMP@PDA@AgNPs. Compared with nanocomposite, smaller inhibition zone was observed for AMP, AgNPs and AMP@PDA. Therefore, there was a synergistic effect between AgNPs and AMP, leading to AMP@PDA@AgNPs with the powerful antibacterial efficacy. The results indicated that the nanocomposite possessed excellent antibacterial property, which can be potentially used as a new antimicrobial agent to effectively treat bacterial infection.
The statistical results of Figure 4A including the diameter and area of the inhibition zone are presented in Figure 4B and C, respectively. As shown in Figure 4B, the diameters of the inhibition zone of the nanocomposite against S. aureus, E. coli and P. aeruginosa were 19 mm, 23 mm and 28 mm, respectively. As shown in Figure 4C, the area of inhibition zone for S. aureus, E. coli and P. aeruginosa with nanocomposite were 1134 mm2, 1661.9 mm2, 2463 mm2, respectively. Compared with the control group, the diameters and area of inhibition zone against S. aureus, E. coli and P. aeruginosa by AMP@PDA@AgNPs were significant difference (**p<0.01, **p <0.001). These results indicated the excellent antibacterial of the nanocomposite against S. aureus, E. coli and P. aeruginosa. It can be clearly seen that the areas of inhibitory zone of nanocomposite against E. coli and P. aeruginosa were larger than that of S. aureus. These results indicated that the nanocomposite had stronger antibacterial effect on Gram-negative bacteria (E. coli, P. aeruginosa) than Gram-positive bacteria (S. aureus), which was attributed to the bacterial structure differences.56 The structural difference in cell walls of bacteria is the combination of biological layer and peptidoglycan layer, phospholipid layer, lipopolysaccharide layer and peptidoglycan layer respectively.57 The researchers found that the AgNPs adhere stably to the bacterial cell wall and penetrate the bacteria, inducing cell death by destroying the cell membrane.58
The MIC is often used as a significant indicator of antibacterial efficacy.59 In this study, the MIC of AMP@PDA@AgNPs against bacteria was determined by the double dilution method. As Figure 5 shows, the growth of all tested bacteria was inhibited with the concentration of 25 μg/mL nanocomposite (Figures 5A–C). In addition, it was found that the number of S. aureus was slightly higher than that of the control group when nanocomposite was 12.5 μg/mL (Figure 5B). These results showed that the nanocomposite had an extremely weak bacteriostatic effect at the concentration of 12.5 μg/mL.
To further assay the antibacterial activity of the AMP@PDA@AgNPs, bacterial activity was examined in LB liquid medium. It could be seen from Figure 5D that the bacterial activity is reduced to 41.48%, 60.68%, 58.99% for E. coli, P. aeruginosa and S. aureus with the presence of AMP@PDA@AgNPs nanocomposite, respectively. However, the bacterial activity was almost 80% under the treatment of AMP, AgNPs and AMP@PDA. The bacterial activity of E. coli, P. aeruginosa and S. aureus treated by AMP@PDA@AgNPs was significant difference from that of PBS control groups (*p<0.05, **p<0.01). These results further demonstrated that the integrating strategy of AMP and AgNPs enhanced the antibacterial performance. According to the above description, the AMP@PDA@AgNPs with enhanced antibacterial ability may be used to destruct biofilms.
Anti-Biofilm Efficacy of AMP@PDA@AgNPs
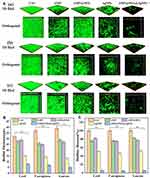
In order to analyze the effect of nanocomposite on biofilm formation, the semi-quantitative analysis of crystal violet staining was applied to quantify the formation of biofilms.60 Interestingly, the result confirmed that the number of bacteria 5×107 CFU/mL was most suitable for the formation of biomass of biofilms (Figure 6A). Compared with the control group, the relative biofilm biomass of S. aureus, E. coli and P. aeruginosa was significant difference at 5×107 CFU/mL (*p<0.05, **p<0.01). As shown in Figure 6A, the biomass of biofilms did not increase as the number of bacteria increased to 1x108 CFU/mL. It was speculated that the growth of biofilms was inhibited due to the limited space of bacteria. As shown in Figure 6B, the biofilms at the bottom of the 96-well plates were slightly removed by treated with AgNPs, AMP and AMP@PDA, whereas significant biofilm removal was seen for the experimental group treated with AMP@PDA@AgNPs. These results suggested that AMP@PDA@AgNPs showed the strongest anti-biofilm effect compared with other groups for all tested bacteria. The quantification of the biofilms in Figure 6B by the semi-quantitative analysis of crystal violet staining is presented in Figure 6C. AMP@PDA@AgNPs showed superior anti-biofilm activity than AgNPs and AMP against both Gram-negative and Gram-positive bacteria (Figure 6C). The biofilm destruction of S. aureus, E. coli and P. aeruginosa by AMP@PDA@AgNPs were significant difference as compared to the control group (*p<0.05, **p<0.01). The result also confirmed that the synergistic antibacterial effect of AMP and AgNPs significantly enhanced the anti-biofilm efficacy of the nanocomposite, which led to the destruction of the biofilms.
To evaluate the effect of nanocomposite on the formation of bacteria biofilms, CLSM was applied to observe the biofilms with different treatment. As shown in the Figure 7A(a), the E. coli biofilms treated with nanocomposite had the weakest fluorescence compared with other groups, indicating that the number of bacteria was remarkably reduced in biofilms. However, the group of AMP and AgNPs, AMP@PDA had stronger fluorescence than AMP@PDA@AgNPs. These results of S. aureus and P. aeruginosa were consistent with the above results (Figure 7A(b) and (c)). The results showed that the combination of the AgNPs and AMP could effectively destroy the bacterial biofilms.
The thickness and biomass of biofilms were quantified by the software ZEN and Image J, respectively. Compared with the PBS control group, the thickness of biofilms in the group of AMP, AMP@PDA and AgNPs, AMP@PDA@AgNPs decreased by 4 µm, 3 µm, 17 µm and 24 µm in E. coli, and 11 µm, 12 µm, 23 µm, 30 µm in P. aeruginosa, 10 µm, 15 µm, 26 µm, 34 µm in S. aureus, respectively (Figure 7B). Compared with the control group, the biofilm thickness of E. coli, P. aeruginosa and S. aureus treated by AMP@PDA@AgNPs was significant difference (*p<0.05, **p<0.01). The most obvious effect was observed for AMP@PDA@AgNPs nanocomposite, the thickness of biofilms reduced down to 8 μm, 7 μm, 5 μm in E. coli, P. aeruginosa, S. aureus, respectively (Figure 7B). The decline of the biomass of biofilms was observed for all groups of AMP, AMP@PDA, AgNPs and AMP@PDA@AgNPs, with the most obvious decline observed for the group of nanocomposite (Figure 7C). Compared with the control group, the biofilm biomass treated with nanocomposite was reduced to 5.99%, 5.23%, 5.74% in E. coli, P. aeruginosa, S. aureus, respectively (Figure 7C). The biofilm biomass of E. coli, P. aeruginosa and S. aureus treated by AMP@PDA@AgNPs was significant difference from that of PBS control groups (*p<0.05, **p<0.01). However, there were still living bacteria under the biofilms. This phenomenon may be ascribed to the stubborn resistance of bacteria to antibiocides. In a word, the AMP@PDA@AgNPs nanocomposite could disrupt and eradicate biofilms of Gram-negative bacteria (P. aeruginosa and E. coli) and Gram-positive bacteria (S. aureus). In this study, AMP@PDA@AgNPs played an important role in destroying the intactness of biofilms and inhibiting the growth of biofilms. And the enhanced anti-biofilm activity of nanocomposite may be ascribed to the synergistic antibacterial effect of AgNPs and AMP.
Inhibitory Effect of AMP@PDA@AgNPs on Biofilm-Related Genes
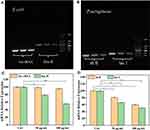
Biofilms are a well-organized bacterial community whose phenotypic growth is related to gene expression and protein production.61 To better understand the suppressed formation of bacterial biofilms with the presence of AMP@PDA@AgNPs, experiments were conducted to reveal the role of gene expression change in the suppression of biofilm formation. After the bacterial biofilms were treated with different concentrations of AMP@PDA@AgNPs, the relative mRNA expression of biofilm-related genes was detected by RT-PCR analysis. The transcription level of las I and rh II, fim H genes was detected in P. aeruginosa, E. coli biofilms with the presence of AMP@PDA@AgNPs (Figure 8).
The transcription level of fim H was decreased with the increased concentrations of the AMP@PDA@AgNPs in E. coli biofilms (Figure 8A). Similarly, the presence of nanocomposite resulted in the reduction of the transcriptional level of las I and rh II in P. aeruginosa biofilms (Figure 8B). The image bands of Figure 8A and B were quantitative analyzed by using Image J software (Figure 8C and D). Under the action of 100 μg/mL nanocomposite, the mRNA relative expression of fim H gene in E. coli decreased to 55.68% compared with the control group (Figure 8C), and the mRNA relative expression of rh II and las I genes decreased to 60.24% and 50.47% in P. aeruginosa, respectively (Figure 8D). The gene expression of las I and rh II, fim H in the AMP@PDA@AgNPs treatment was a significant difference as compared to the control group (*p<0.05, **p<0.01). The result confirmed that the mRNA expression of biofilm-related genes was inhibited by the nanocomposite. More importantly, it was speculated that the AMP@PDA@AgNPs prevented the formation of biofilms by decreasing the protein expression of las I and rh II, fim H. The exploration of antibacterial mechanism indicated that the nanocomposite may preclude the formation of biofilm by inhibiting the transcription level of biofilm-related genes. The specific mechanism of nanoparticle-mediated antibacterial activity is still unclear. Metal nanoparticles interact with microorganisms through a variety of mechanisms which comprise the enzyme degradation, inactivation of major cellular proteins and impairment of genetic materials, generation of reactive oxygen species (ROS).62 Siemer et al investigate that nanoparticles invade bacterial cells by changing the phospholipid composition of the cell membrane to adjust the surface charge.63 Exploring more antimicrobial mechanisms of nanomaterials will help us more effectively eliminate bacterial infections in the future.
Conclusion
In summary, the AMP@PDA@AgNPs nanocomposite with good biocompatibility was successfully prepared. The nanocomposite could destruct bacterial biofilms and inhibit bacterial growth by inhibiting the expression of biofilm-related genes. The synergistic strategy of AMPs and AgNPs could provide a new perspective for the treatment of bacterial infection. It shows a great application potential in the field of clinic infection.
Acknowledgments
This work was financially supported by Key Science and Technology Program of Henan Province (192102310189) and Innovation Scientists and Technicians Troop Construction Projects of Henan Province (CXTD2017071).
Disclosure
The authors declare no conflict of interest.
References
1. Lu L, Hu W, Tian Z, et al. Developing natural products as potential anti-biofilm agents. Chin Med. 2019;14. doi:10.1186/s13020-019-0232-2
2. Medina-Rodriguez AC, Avila-Sierra A, Ariza JJ, et al. Clean-in-place disinfection of dual-species biofilm (Listeria and Pseudomonas) by a green antibacterial product made from citrus extract. Food Control. 2020;118:107422. doi:10.1016/j.foodcont.2020.107422
3. Zhang C, Du C, Liao J-Y, et al. Synthesis of magnetite hybrid nanocomplexes to eliminate bacteria and enhance biofilm disruption. Biomater Sci. 2019;7(7):2833–2840. doi:10.1039/c9bm00057g
4. Kathirvel B, Felix L, Eleftherios M, Sabarathinam S, Nath VT, Arivalagan P. Biofilm and Quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020.
5. Lilit G, Hamlet B, Vladimir G, Armen T. Comparable antibacterial effects and action mechanisms of silver and iron oxide nanoparticles on Escherichia coli and Salmonella typhimurium. Sci Rep. 2020;10(1).
6. Han W, Zhou B, Yang K, et al. Biofilm-inspired adhesive and antibacterial hydrogel with tough tissue integration performance for sealing hemostasis and wound healing. Bioact Mater. 2020;5(4):768–778. doi:10.1016/j.bioactmat.2020.05.008
7. Yifan G, Jing W, Mengying C, et al. Size and charge adaptive clustered nanoparticles targeting the biofilm microenvironment for chronic lung infection management. ACS Nano. 2020;14(5).
8. Elshaarawy RFM, Ismail LA, Alfaifi MY, Rizk MA, Eltamany EE, Janiak C. Inhibitory activity of biofunctionalized silver-capped N-methylated water-soluble chitosan thiomer for microbial and biofilm infections. Int J Biol Macromol. 2020;152:709–717. doi:10.1016/j.ijbiomac.2020.02.284
9. Riau AK, Aung TT, Setiawan M, et al. Surface immobilization of nano-silver on polymeric medical devices to prevent bacterial biofilm formation. Pathogens. 2019;8(3):93. doi:10.3390/pathogens8030093
10. Dokoshi T, Zhang LJ, Li F, et al. Hyaluronan degradation by cemip regulates host defense against Staphylococcus aureus skin infection. Cell Rep. 2020;30(1):61–68.e4. doi:10.1016/j.celrep.2019.12.001
11. Mala R, Aglin AA, Celsia ASR, et al. Foley catheters functionalised with a synergistic combination of antibiotics and silver nanoparticles resist biofilm formation. Iet Nanobiotechnol. 2017;11(5):612–620. doi:10.1049/iet-nbt.2016.0148
12. Zhu J, Wang J, Hou J, Zhang Y, Liu J, Van der Bruggen B. Graphene-based antimicrobial polymeric membranes: a review. J Mater Chem A. 2017;5(15):6776–6793. doi:10.1039/c7ta00009j
13. Guo J, Qin S, Wei Y, et al. Silver nanoparticles exert concentration-dependent influences on biofilm development and architecture. Cell Prolif. 2019;52(4). doi:10.1111/cpr.12616
14. Vanaraj S, Keerthana BB, Preethi K. Biosynthesis, characterization of silver nanoparticles using quercetin from clitoria ternatea L to enhance toxicity against bacterial biofilm. J Inorg Organomet Polym Mater. 2017;27(5):1412–1422. doi:10.1007/s10904-017-0595-8
15. Yuan Z, Lin C, He Y, et al. Near-infrared light-triggered nitric-oxide-enhanced photodynamic therapy and low-temperature photothermal therapy for biofilm elimination. Acs Nano. 2020;14(3):3546–3562. doi:10.1021/acsnano.9b09871
16. Fan W, Liu D, Li Y, Sun Q, Fan B. AgCa-PLGA submicron particles inhibit the growth and colonization of E. Faecalis and P. Gingivalis on dentin through infiltration into dentinal tubules. Int J Pharm. 2018;552(1–2):206–216. doi:10.1016/j.ijpharm.2018.09.066
17. Mohamed AA, Abu-Elghait M, Ahmed NE, Salem SS. Eco-friendly mycogenic synthesis of ZnO and CuO nanoparticles for in vitro antibacterial, antibiofilm, and antifungal applications. Biol Trace Elem Res. 2021;199(7):2788–2799. doi:10.1007/s12011-020-02369-4
18. Mori AD, Jones RS, Cretella M, et al. Evaluation of antibacterial and cytotoxicity properties of silver nanowires and their composites with carbon nanotubes for biomedical applications. Int J Mol Sci. 2020;21(7).
19. Sheng Z, Van Nostrand JD, Zhou J, Liu Y. Contradictory effects of silver nanoparticles on activated sludge wastewater treatment. J Hazard Mater. 2018;341:448–456. doi:10.1016/j.jhazmat.2017.07.051
20. Pounraj S, Somu P, Paul S. Chitosan and graphene oxide hybrid nanocomposite film doped with silver nanoparticles efficiently prevents biofouling. Appl Surf Sci. 2018;452:487–497. doi:10.1016/j.apsusc.2018.05.009
21. Namivandi-Zangeneh R, Wong EH, Boyer C. Synthetic antimicrobial polymers in combination therapy: tackling antibiotic resistance. ACS Infect Dis. 2021;7(2):215–253. doi:10.1021/acsinfecdis.0c00635
22. Dharshini KP, Dominik B, Devi DR, et al. Silver nanomaterials for wound dressing applications. Pharmaceutics. 2020;12(9).
23. Dutra-Correa M, Leite AABV, de Cara SPHM, et al. Antibacterial effects and cytotoxicity of an adhesive containing low concentration of silver nanoparticles. J Dent. 2018;77:66–71. doi:10.1016/j.jdent.2018.07.010
24. Enan ET, Ashour AA, Basha S, Felemban NH, Gad El-Rab SMF. Antimicrobial activity of biosynthesized silver nanoparticles, amoxicillin, and glass-ionomer cement against Streptococcus mutans and Staphylococcus aureus. Nanotechnology. 2021;32(21):215101. doi:10.1088/1361-6528/abe577
25. Rodrigues AG, RdC R, Selari PJRG, de Araujo WL, de Souza AO. Anti-biofilm action of biological silver nanoparticles produced by aspergillus tubingensis and antimicrobial activity of fabrics carrying it. Biointerface Res Appl Chem. 2021;11(6):14764–14774. doi:10.33263/briac116.1476414774
26. Shri RK, Subitha P, Narasimhan S, Murugesan R, Narayan S. Fabrication of dexamethasone-silver nanoparticles entrapped dendrimer collagen matrix nanoparticles for dental applications. Biointerface Res Appl Chem. 2021;11(6):14935–14955. doi:10.33263/briac116.1493514955
27. Gao J, Na H, Zhong R, et al. One step synthesis of antimicrobial peptide protected silver nanoparticles: the core-shell mutual enhancement of antibacterial activity. Colloids Surf B Biointerfaces. 2020;186:110704. doi:10.1016/j.colsurfb.2019.110704
28. Felipe Salas-Orozco M, Nino Martinez N, Martinez-Castanon GA, Torres Mendez F, Patino-Marin N, Ruiz F. Detection of genes related to resistance to silver nanoparticles in bacteria from secondary endodontic infections. J Nanomater. 2019;2019. doi:10.1155/2019/8742975
29. Kawai R, Niki M, Yada S, Yoshimura T. Physicochemical and solution properties of quaternary-ammonium-salt-type amphiphilic gemini ionic liquids with spacers containing oxygen or nitrogen. Colloids Surf a Physicochem Eng Asp. 2020;603. doi:10.1016/j.colsurfa.2020.125218
30. Prabhawathi V, Sivakumar PM, Boobalan T, Manohar CM, Doble M. Design of antimicrobial polycaprolactam nanocomposite by immobilizing subtilisin conjugated Au/Ag core-shell nanoparticles for biomedical applications. Mater Sci Eng C. 2019;94:656–665. doi:10.1016/j.msec.2018.10.020
31. Saafan A, Zaazou MH, Sallam MK, Mosallam O, El Danaf HA. Assessment of photodynamic therapy and nanoparticles effects on caries models. Open Access Maced J Med Sc. 2018;6(7):1289–1295. doi:10.3889/oamjms.2018.241
32. Parasuraman P, R. y T, Shaji C, et al. Biogenic silver nanoparticles decorated with methylene blue potentiated the photodynamic inactivation of Pseudomonas aeruginosa and Staphylococcus aureus. Pharmaceutics. 2020;12(8):709. doi:10.3390/pharmaceutics12080709
33. Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother. 2010;54(8):3132–3142. doi:10.1128/aac.00124-10
34. Schnaider L, Brahmachari S, Schmidt NW, et al. Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. Nat Commun. 2017;8. doi:10.1038/s41467-017-01447-x
35. Lu Z, Wu Y, Cong Z, et al. Effective and biocompatible antibacterial surfaces via facile synthesis and surface modification of peptide polymers. Bioact Mater. 2021;6(12):4531–4541. doi:10.1016/j.bioactmat.2021.05.008
36. Ageitos JM, Sanchez-Perez A, Calo-Mata P, Villa TG. Antimicrobial peptides (AMPs): ancient compounds that represent novel weapons in the fight against bacteria. Biochem Pharmacol. 2017;133:117–138. doi:10.1016/j.bcp.2016.09.018
37. Yajuan S, Luca MV, Hongjun W, et al. Dissolvable microneedles coupled with nanofiber dressings eradicate biofilms via effectively delivering a database-designed antimicrobial peptide. ACS Nano. 2020.
38. Cheng W, Zeng X, Chen H, et al. Versatile polydopamine platforms: synthesis and promising applications for surface modification and advanced nanomedicine. Acs Nano. 2019;13(8):8537–8565. doi:10.1021/acsnano.9b04436
39. Sun Y, Deng Y, Ye Z, Liang S, Tang Z, Wei S. Peptide decorated nano-hydroxyapatite with enhanced bioactivity and osteogenic differentiation via polydopamine coating. Colloids Surf B Biointerfaces. 2013;111:107–116. doi:10.1016/j.colsurfb.2013.05.037
40. Zhang M, Zou Y, Zuo C, et al. Targeted antitumor comparison study between dopamine self-polymerization and traditional synthesis for nanoparticle surface modification in drug delivery. Nanotechnology. 2021;32(30):305102. doi:10.1088/1361-6528/abf8dd
41. Tong C, Zhong X, Yang Y, et al. PB@PDA@Ag nanosystem for synergistically eradicating MRSA and accelerating diabetic wound healing assisted with laser irradiation. Biomaterials. 2020;243:119936. doi:10.1016/j.biomaterials.2020.119936
42. Dong PV, Ha CH, Binh LT, Kasbohm J. Chemical synthesis and antibacterial activity of novel-shaped silver nanoparticles. Int Nano Lett. 2012;2(1).
43. Shuai C, Xu Y, Feng P, Wang G, Xiong S, Peng S. Antibacterial polymer scaffold based on mesoporous bioactive glass loaded with in situ grown silver. Chem Eng J. 2019;374:304–315. doi:10.1016/j.cej.2019.03.273
44. Yuwen LH, Sun YT, Tan GL, et al. MoS2@polydopamine-Ag nanosheets with enhanced antibacterial activity for effective treatment of Staphylococcus aureus biofilms and wound infection. Nanoscale. 2018;10(35):16711–16720. doi:10.1039/c8nr04111c
45. Ansari MA, Asiri SMM, Alzohairy MA, Alomary MN, Almatroudi A, Khan FA. Biofabricated fatty acids-capped silver nanoparticles as potential antibacterial, antifungal, antibiofilm and anticancer agents. Pharmaceuticals. 2021;14(2):139. doi:10.3390/ph14020139
46. Sun YH, Qin HS, Yan ZQ, Zhao CQ, Ren JS, Qu XG. Combating biofilm associated infection in vivo: integration of quorum sensing inhibition and photodynamic treatment based on multidrug delivered hollow carbon nitride sphere. Adv Funct Mater. 2019;29(14). doi:10.1002/adfm.201808222
47. Katani R, Kudva IT, Srinivasan S, et al. Strain and host-cell dependent role of type-1 fimbriae in the adherence phenotype of super-shed Escherichia coli O157: H7. Int J Med Microbiol. 2021;311(4):151511. doi:10.1016/j.ijmm.2021.151511
48. Erdal B, Yalinay M, Elmas C, Yazici GN. Investigation of pseudomonas aeruginosa biofilm formation and quorum sensing genes in piperacillin/tazobactam and ciprofloxacin sub-minimal inhibitory concentrations. Mikrobiyol Bul. 2020;54(4):547–558. doi:10.5578/mb.70087
49. Guo Q, Zhao Y, Dai X, et al. Functional silver nanocomposites as broad-spectrum antimicrobial and biofilm-disrupting agents. ACS Appl Mater Interfaces. 2017;9(20):16835–16848. doi:10.1021/acsami.7b02775
50. Zheng Y, Liu W, Chen Y, Li C, Jiang H, Wang X. Conjugating gold nanoclusters and antimicrobial peptides: from aggregation-induced emission to antibacterial synergy. J Colloid Interface Sci. 2019;546:1–10. doi:10.1016/j.jcis.2019.03.052
51. Pal I, Brahmkhatri VP, Bera S, et al. Enhanced stability and activity of an antimicrobial peptide in conjugation with silver nanoparticle. J Colloid Interface Sci. 2016;483:385–393. doi:10.1016/j.jcis.2016.08.043
52. Joshi S, Siddiqui R, Sharma P, Kumar R, Verma G, Saini A. Green synthesis of peptide functionalized reduced graphene oxide (rGO) nano bioconjugate with enhanced antibacterial activity. Sci Rep. 2020;10(1). doi:10.1038/s41598-020-66230-3
53. Mohanta YK, Panda SK, Jayabalan R, Sharma N, Bastia AK, Mohanta TK. Antimicrobial, antioxidant and cytotoxic activity of silver nanoparticles synthesized by leaf extract of Erythrina suberosa (Roxb.). Front Mol Biosci. 2017;4. doi:10.3389/fmolb.2017.00014
54. Zhang X, Zhang G, Chai M, Yao X, Chen W, Chu PK. Synergistic antibacterial activity of physical-chemical multi-mechanism by TiO2 nanorod arrays for safe biofilm eradication on implant. Bioact Mater. 2021;6(1):12–25. doi:10.1016/j.bioactmat.2020.07.017
55. Liang G, Shi H, Qi Y, et al. Specific anti-biofilm activity of carbon quantum dots by destroying P. gingivalis biofilm related genes. Int J Nanomedicine. 2020;Volume 15:5473–5489. doi:10.2147/IJN.S253416
56. El-Fiqi A, Mandakhbayar N, Jo SB, Knowles JC, Lee JH, Kim HW. Nanotherapeutics for regeneration of degenerated tissue infected by bacteria through the multiple delivery of bioactive ions and growth factor with antibacterial/angiogenic and osteogenic/odontogenic capacity. Bioact Mater. 2021;6(1):123–136. doi:10.1016/j.bioactmat.2020.07.010
57. Alomary MN, Ansari MA. Proanthocyanin-capped biogenic TiO2 nanoparticles with enhanced penetration, antibacterial and ROS mediated inhibition of bacteria proliferation and biofilm formation: a comparative approach. Chem – A Eur J. 2021;27(18):5817–5829. doi:10.1002/chem.202004828
58. Kishore MY, Kunal B, Kumar JS, Abeer H, Fathi AAE, Kumar MT. Anti-biofilm and antibacterial activities of silver nanoparticles synthesized by the reducing activity of phytoconstituents present in the Indian medicinal plants. Front Microbiol. 2020;11.
59. Wang Y, Xiao F, Jin G. Structural basis of quinolone derivatives, inhibition of type I and II topoisomerases and inquiry into the relevance of bioactivity in odd or even branches with molecular docking study. J Mol Struct. 2020;1221:128869. doi:10.1016/j.molstruc.2020.128869
60. Arafa MG, Mousa HA, Afifi NN. Preparation of PLGA-chitosan based nanocarriers for enhancing antibacterial effect of ciprofloxacin in root canal infection. Drug Deliv. 2020;27(1):26–39. doi:10.1080/10717544.2019.1701140
61. Hu DF, Deng YY, Jia F, Jin Q, Ji J. Surface charge switchable supramolecular nanocarriers for nitric oxide synergistic photodynamic eradication of biofilms. Acs Nano. 2020;14(1):347–359. doi:10.1021/acsnano.9b05493
62. Mohanta YK, Hashem A, Abd_Allah EF, Jena SK, Mohanta TK. Bacterial synthesized metal and metal salt nanoparticles in biomedical applications: an up and coming approach. Appl Organomet Chem. 2020;34(9). doi:10.1002/aoc.5810
63. Siemer S, Westmeier D, Barz M, et al. Biomolecule-corona formation confers resistance of bacteria to nanoparticle-induced killing: implications for the design of improved nanoantibiotics. Biomaterials. 2019;192:551–559. doi:10.1016/j.biomaterials.2018.11.028
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.