Back to Journals » ImmunoTargets and Therapy » Volume 4
Emerging roles for HMGB1 protein in immunity, inflammation, and cancer
Authors Martinotti S, Patrone M, Ranzato E
Received 7 March 2015
Accepted for publication 14 April 2015
Published 26 May 2015 Volume 2015:4 Pages 101—109
DOI https://doi.org/10.2147/ITT.S58064
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 7
Editor who approved publication: Professor Michael Shurin
Simona Martinotti, Mauro Patrone, Elia Ranzato
DiSIT – Dipartimento di Scienze e Innovazione Tecnologica, University of Piemonte Orientale, Alessandria, Italy
Abstract: High-mobility group box 1 (HMGB1) protein is a member of the highly conserved non-histone DNA binding protein family. First identified in 1973, as one of a group of chromatin-associated proteins with high acidic and basic amino acid content, it was so named for its characteristic rapid mobility in polyacrylamide gel electrophoresis. HMGB1 was later discovered to have another function. It is released from a variety of cells into the extracellular milieu to act on specific cell-surface receptors. In this latter role, HMGB1 is a proinflammatory cytokine that may contribute to many inflammatory diseases, including sepsis. Therefore, HMGB1 regulates intracellular cascades influencing immune cell functions, including chemotaxis and immune modulation. The bioactivity of the HMGB1 is determined by specific posttranslational modifications that regulate its role in inflammation and immunity. During tumor development, HMGB1 has been reported to play paradoxical roles in promoting both cell survival and death by regulating multiple signaling pathways. In this review, we focus on the role of HMGB1 in physiological and pathological responses, as well as the mechanisms by which it contributes to immunity, inflammation, and cancer progression.
Keywords: alarmin, DNA replication, HMGB1, posttranslational modifications, wound repair
Introduction
The high-mobility group box 1 (HMGB1), a member of the high-mobility group (HMG) protein family, was identified in calf thymus in 1973 by Goodwin et al.1 The name “HMG” is derived from its high electrophoretic mobility in polyacrylamide gels.
HMG proteins are subdivided into three families named HMGB, HMGA, and HMGN. All HMGB proteins, including HMGB1–4, contain the functional HMG-box motif, have architectural functions, and are capable of organizing dynamic active chromatin structures. In adults, HMGB1 is expressed everywhere except in neurons, whereas HMGB2 and HMGB4 are expressed in the testes, and HMGB3 primarily in lymphoid organs.2 Prior to its naming, HMGB1 was also known as HMG1, amphoterin, and SBP-1.3
HMGB1 senses and coordinates the cellular stress response and plays a critical role not only inside of the cell as a DNA chaperone, chromosome guardian, autophagy sustainer, and protector from apoptotic cell death, but also outside the cell as the prototypic damage-associated molecular pattern molecule (DAMP).4
Therefore, to elucidate the pleiotropic roles of HMGB1, in this review, we will emphasize the characteristics that make HMGB1 a critical molecular target in diseases including infectious diseases, ischemia, immune disorders, neurodegenerative diseases, metabolic disorders, and cancer.
HMGB1: structural characteristics
HMGB1 is expressed in all vertebrate cells, in yeast, plants, and bacteria, localized in cytoplasm and nuclei. The protein abundance is high in thymus (106 molecules/cell) and in undifferentiated tissues; it is highly conserved through evolution, and has 99% identity among all mammals. It probably originated more than 500 million years ago, before the split between the animal and plant kingdoms.5
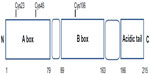
HMGB1 structure is composed of two tandem DNA-binding HMG-box domains (N-terminal A and central B) and an acidic C-terminal tail, mediating protein–protein interactions (Figure 1).
HMGB1 is a 215 amino acid protein of 30 kDa, composed of three domains: two positively charged domains (A box and B box) and a negatively charged carboxyl terminus (acidic tail). This structure confers onto HMGB1 the peculiar feature to recognize and specifically bind DNA structures, containing sharp bends or kinks, such as four-way junctions.6,7
Structurally, HMGB1 consists of 215 residues organized in three main functional domains: the A and B boxes (positively charged) and the acidic tail (negatively charged). The A and B boxes, residues 1–79 and 89–163, respectively, are functionally DNA-binding domains.8,9
This region is composed of α-helical structures; in the nucleus, HMGB1 binds to the minor groove of linear DNA and bends it into a helical structure.10 The C-terminal acidic tail, residues 186–215, plays an important role in nuclear functions.11,12
At the N-terminus of HMGB1 (residues 6–12) is a heparin-binding sequence that likely contributes to the heparin and heparin sulfate binding capacity of HMGB1.12
A C-terminal region of HMGB1 at amino acids 150–183 mediates the receptor for advanced glycation end-product (RAGE) and Toll-like receptor (TLR) binding.13
The RAGE is a cell-bound receptor of the immunoglobulin superfamily that is activated by a variety of proinflammatory ligands, including HMGB1, advanced glycation end-products, members of the S100 family of proteins, and amyloid β-peptide.14
Three cysteines (Cys) residues, crucial for the biological activities of extracellular HMGB1, are encoded within the HMGB1: two vicinal Cys in box A (C23 and C45) and a single one in box B (C106).15,16
Extracellular HMGB1 can bind and activate different signaling transduction cell receptors, such as the receptor RAGE; the TLRs 2, 4, and 9; macrophage antigen-1; syndecan-3; CD24-Siglec-10; CXCR4; and certain integrin.17
RAGE is expressed in endothelial cells, vascular smooth muscle cells, monocytes, macrophages, dendritic cells, neurons, and glial cells.18 The interaction of RAGE–HMGB1 leads to the activation of different signal transduction pathways and culminates in cell responses ranging from cell motility to cell proliferation, as well as the production and release of cytokines/chemokines. RAGE blockade suppresses the HMGB1 effects, but only partially, strongly suggesting the existence of additional receptors for HMGB1. TLR2 and 4 are involved in HMGB1-induced proinflammatory activation of macrophages and neutrophils. TLRs blockade does not completely abolish HMGB1 effect.
Recently, new receptor-mediating HMGB1 effects were demonstrated;19 these authors evaluated that at concentrations of agonist that were per se ineffective, HMGB1 potentiates the activation of the ionotropic glutamate N-methyl-d-aspartate receptor, interacting with the ion channel through the sequence corresponding to the peptide located in the box B at the amino acids 130–139.9
Known roles in DNA replication, DNA repair, and transcription
HMGB1 is a member of the HMG family whose nuclear localization and affinity for DNA is regulated through phosphorylation and acetylation. HMGB1 has a dynamic relationship with chromatin and plays an important role in DNA architecture and transcriptional regulation.20
HMGB1 is stored in the nucleus as a result of the presence of two lysine-rich nuclear localization sequences (NLSs) located in the box A (residues 28–44) and in the box B (residues 179–185). Hyperacetylation of NLSs promotes HMGB1 translocation from the nucleus to the cytosol, and the subsequent release of HMGB1.21
The subcellular localization of HMGB1 depends on the type and activation state of the cell. The HMGB1 is usually localized in the cell nucleus and diffusely distributed in cytoplasm.22 HMGB1 can be released from the nucleus to the extracellular space in response to different stimuli in two ways: following cellular injury it is passively released during cellular apoptosis or necrosis; or it is released actively following inflammatory signals from activated immune cells or neuronal cells. Hyperacetylated HMGB1 molecules accumulate in the cytosol where they are packaged into secretory lysosomes and then released into the extracellular environment.23
HMGB1 binds the minor groove of DNA facilitating the assembly of site-specific DNA-binding proteins, including nuclear hormone/nuclear hormone receptor complexes and p53 or p73 transcriptional complexes.24
HMGB1 is involved in the facilitation of protein–protein interaction and recognition of DNA damage in the process of mismatch repair.25
HMGB1 is the structural protein of chromatin and regulates nuclear homeostasis and genome stability in several ways: HMGB1 binds to nucleosomes at the dyad axis, promotes nucleosome sliding, relaxes nucleosome structure, and makes chromatin more accessible by its ability to bend DNA.26
HMGB1 has been found to increase the binding affinity of many sequence-specific transcription factors to their cognate DNA, such as p53, p73, the retinoblastoma (Rb) protein, nuclear factor-κB (NF-κB), and the estrogen receptor.
HMGB1 directly binds to a variety of bulky DNA lesions which allows it to participate in DNA repair pathways including nucleotide excision repair, base excision repair, mismatch repair, and double strand break repair via nonhomologous end-joining.
Loss of HMGB1 reduces telomerase activity, decreases telomere length, and increases chromosomal instability.27
HMGB1 as an alarmin: roles in inflammation, response to infection, and wound repair
For HMGB1 to function as a cytokine, it must be released into the extracellular milieu. HMGB1 is released passively during cellular necrosis by almost all cells that have a nucleus and signals neighboring cells of ongoing damage.
In 1999, Wang et al first reported that HMGB1 was liberated from cells stimulated with cytokines and that HMGB1 played an important role in mediating experimental sepsis.28 Then, Scaffidi et al29 showed that HMGB1 is released from cells undergoing necrosis, but not from apoptotic cells, because HMGB1 is tightly bound to chromatin.
Recent reports demonstrate that HMGB1 can also be released by apoptotic cells,30 but in this case, the released HMGB1 appears to have a tolerogenic rather than a proinflammatory effect.31
Active secretion of HMGB1 is generally by nontraditional, leaderless pathways which are not routed through the endoplasmic reticulum or Golgi apparatus. During normal cellular homeostasis the dynamic relationship of HMGB1 with the nucleus and cytoplasm heavily favors the nucleus. However, HMGB1 concentrates in secretory lysosomes when hyperacetylated on lysine residues and then is released upon appropriate signaling stimuli.28
When HMGB1 is not acetylated, it remains localized to the nucleus and is not secreted or released, such as during apoptosis.31
Because HMGB1 does not contain a leader sequence, it is released via a nonclassical secretory pathway that may involve specialized vesicles of the endolysosomal compartment. Stimuli for secretion of HMGB1 are diverse and include pathogen-associated molecular patterns, cytokines, and certain states of cellular stress.
HMGB1 can promote inflammatory responses by numerous mechanisms. Comparisons of the necrotic cell debris from HMGB1-deficient and wild-type cells demonstrate that HMGB1-deficient cells have a profoundly reduced capacity to induce cytokines.29
Recombinant HMGB1 also induces cytokine production from human monocytes but not from lymphocytes.32 However, several groups have more recently shown that highly purified HMGB1 does not induce significant amounts of proinflammatory cytokines.6
However, it should also be noted that recombinant HMGB1 may be different from the native protein, with changes in the oxidative status having a profound impact on its biological activity.31
HMGB1 is not released only from cells of the immune system; for other type of cells, it has been demonstrated an active secretion pathway. The first nonimmune cell to be studied for active HMGB1 secretion was the pituicyte, which was found to release HMGB1 in response to IL-1 or TNF stimulation.28
Sepsis is a lethal syndrome after infection or injury, associated with a high mortality rate,33 and proinflammatory cytokines released from monocyte/macrophages and other cells have been implicated as important mediators of the lethal effect of endotoxin.28 Proinflammatory cytokines can lead to the development of tissue damage, metabolic acidosis, hypotension, multiple organ failure, and even death.33
Endogenous danger signals released from necrotic or stressed cells that trigger the inflammatory response after trauma have been termed alarmins or danger-associated molecular patterns (DAMPs).
HMGB1 was recognized as a late mediator of sepsis in animal models of systemic endotoxemia.28 Therapy with anti-HMGB1 antibodies before or after endotoxin exposure confers significant protection against lethality, and administration of HMGB1 itself was lethal, suggesting that HMGB1 is an endogenous mediator of endotoxin lethality.28
Some research reported that cytokines may stimulate release of HMGB1 from pituicytes and astrocytes.34 These findings suggest a role for HMGB1 in neurological disorders. It also demonstrates that HMGB1 is highly expressed in mouse brain neurons and astrocytes, is released during brain ischemia, and contributes to neuroinflammation and postischemic brain damage.35
Clinical studies have demonstrated that HMGB1 is a late mediator of sepsis and some investigations have reported elevated serum/plasma HMGB1 concentrations in patients with sepsis.28
Until now, it was not known to what extent local HMGB1 release contributes to serum HMGB1 levels, and to what extent it contributes to damage at the site of infection and to distant organs in patients with sepsis.33 It has been reported that genetic variation in the HMGB1 gene could affect the outcome of patients with systemic inflammatory response syndrome and sepsis, suggesting a possible role for HMGB1 genetics.36
More recently, HMGB1 has been associated, as a putative danger signal, to pathogenesis of a variety of noninfectious inflammatory conditions, ie, autoimmunity, trauma, hemorrhagic shock, and ischemia-reperfusion injury.37 Furthermore, a role for HMGB1 has been suggested in some normal and pathological conditions for a huge number of organs and systems including liver, heart, pancreas, brain, bone, and kidney.38 In fact, HMGB1 levels are elevated in patients with mechanical trauma, such as strokes, acute myocardial infarction, acute respiratory distress, and liver transplantation.39,40 This pleiotropic inflammatory role for HMGB1 could be due to the existence of multiple receptors that mediate HMGB1 responses. The different responses to HMGB1 stimulation may rely on the levels of HMGB1, on the expression of the receptors in the target organ, and on the ability of HMGB1 to interact with other molecules. Potentially, there is significant cross talk between receptors, and the deficient or defective receptor may influence the function of other receptors.41
There is growing interest in the role of alarmins, such as HMGB1, in contrast to their role in promoting inflammation, to promote tissue repair and regeneration.42 HMGB1 may act as a trigger of tissue repair, recruiting stem cells and promoting their proliferation.43
The physiological meaning of the role of HMGB1 in wound healing remains to be elucidated. For cutaneous wound healing, some studies42,44 allowed us to postulate that HMGB1 release may occur from skin cells as the result of cell activation, enhanced membrane permeability, or membrane ruptures. After its release in the wounded area, HMGB1 could operate the recruitment of different cell types, influencing keratinocyte and fibroblast proliferation and migration.42
Taken together, the regenerative properties of HMGB1 represent a potentially important way by which the functions of such alarmin can be manipulated to promote healing as opposed to injury.
Emerging roles in innate and adaptive immunity and autoimmunity
It has also been found that HMGB1 behaves as an endogenous immune adjuvant.45 HMGB1 elicits a striking increase in the titers of high-affinity hypermutated IgG when coinjected with the soluble nominal antigen ovalbumin. Moreover, it was sufficient to reconstitute the immunogenicity of apoptotic “self” cells in vivo.46
HMGB1 triggers inflammation, attracting other cells, inducing tissue repair, recruiting stem cells, and promoting their proliferation. HMGB1 also activates dendritic cells and promotes their functional maturation and their response to lymph node chemokines. Therefore, HMGB1 acts in an autocrine/paracrine fashion and sustains long-term repair and defense programs. HMGB1 secretion is critical for the immunity system because dendritic cells, when reaching the lymph nodes, secrete HMGB1, sustaining the proliferation of antigen-specific T-cells, to prevent their activation-dependent apoptosis, and to promote their polarization toward a T-helper 1 phenotype.46
Considerable evidence suggests the implication of extracellular HMGB1 in the pathogenesis of a variety of autoimmune diseases.
Anti-HMGB1 antibodies are present in the serum of patients with rheumatoid arthritis and drug-induced systemic lupus erythematosus.47 HMGB1 is overexpressed in synovial biopsy specimens of rheumatoid patients and in experimental arthritis.48
HMGB1 induces arthritis in mice, while treatment with HMGB1 antagonists ameliorates collagen-induced arthritis in both rats and mice.49
Increased extracellular HMGB1 is also detectable in the dermis and epidermis of skin lesions in patients with cutaneous lupus erythematosus and in biopsy specimens of the minor salivary glands of patients with Sjogren’s syndrome.50
In multiple sclerosis, HMGB1 expression was found to be upregulated in brain active lesions from patients.51 In the animal model of multiple sclerosis, experimental autoimmune encephalomyelitis, neutralization of HMGB1 ameliorated clinical severity, reduced central nervous system pathology, and blocked proinflammatory cytokine production.52
New recent data support a novel concept of brain–immune interaction after acute brain lesions in which soluble danger signals from the brain mediate a complex immune-modulatory reaction in the peripheral immune system.53,54 The HMGB1-RAGE-mediated pathway could be a key mechanism explaining the complex postischemic brain–immune interactions. This mechanism indicates that the cytokine-inducing, fully reduced isoform of HMGB1 was released from the ischemic brain in the hyperacute phase of stroke in mice and patients. Cytokines secreted in the periphery in response to brain injury induced sickness behavior, which could be abrogated by inhibition of the HMGB1-RAGE pathway or direct cytokine neutralization.54 Therefore, HMGB1 in the central nervous system could a useful biomarker and may contribute to the chronicity of neuroinflammation through the proposed positive-feedback loop involving infiltrating macrophages and resident microglia.51
Posttranslational modifications and molecular interactions in HMGB1 function
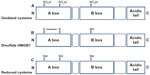
HMGB1 contains three Cys residues, which are critical for the biological activity of the protein (Figure 2). Cys are modified by different redox signals with oxidation of thiol group side-chains (-SH) to several reversible redox states, such as disulfide (R-S-S-R), sulfenic acid (R-SOH), and sulfonate (R-SO3H) moieties.37
HMGB1 can form a Cys23–Cys45 intermolecular disulfide bond, while the Cys106 remains in the reduced form.15
Replacement of Cys23 and/or Cys45 with serines did not affect the nuclear distribution of the mutant proteins, whereas C106 and triple Cys mutations impaired nuclear localization of HMGB1.55
The redox status of HMGB1 recognized its cytokine-inducing and chemokine activity.16 C106 is necessary for the binding of HMGB1 to TLR4 to stimulate cytokine release and inflammation.56
Recent observations suggested that the disulfide bond between C23 and C45 is also required for HMGB1 cytokine activity.16 Indeed, mutations of C45 or C23 abolish HMGB1 cytokine-inducing activity.
Reactive oxygen species oxidizes HMGB1 at C106 released from apoptotic cells, thereby neutralizing its cytokine potential.31 All-cysteine-oxidized HMGB1 conformation prevents HMGB1’s cytokine or chemokine activity.16 Therefore, the redox modifications of HMGB1 play a crucial role for the protein functionality during infection and inflammation.56
The complex and fine-tuned process of autophagy is also regulated by HMGB1 activities. During stress, starvation, oxidative stress, and chemotherapy, HMGB1 translocate from nucleus to cytoplasm for binding beclin-1, which initiates the autophagosome process. The intramolecular disulfide bond (C23–C45) of HMGB1 is required for binding beclin-1. Once released, this reduced-HMGB1 triggers autophagy in a RAGE-dependent manner in cancer cells.56
The functional in vivo relevance of redox-modified HMGB1 has been confirmed in experimental animal models.20
HMGB1 may undergo other extensive posttranslational modifications, including reversible acetylation, methylation, ADP ribosylation, glycation, and phosphorylation.57
Acetylation of key lysine residues within the two NLS sites of HMGB1 is a regulatory mechanism for intracellular shuttling, allowing the release of protein from activated monocytes and macrophages.58 Hyperacetylation of these residues unbalances the HMGB1 from its nuclear position toward cytoplasmic accumulation and its continuous shuttling between nucleus and cytoplasm.59 The acetylated HMGB1 in serum of patients is a specific biomarker of acetaminophen-induced hepatotoxicity that is useful for clinical outcome.60 Other findings suggest that the hyperacetylated HMGB1 is a newly identified biomarker for pyroptosis. Pyroptosis is a proinflammatory cell-death mode associated with osmotic lysis and the release of the intracellular content into the extracellular milieu.61 Redox status of three Cys of HMGB1 suggest that HMGB1 released during pyroptosis may contain active (disulfide-bonded) form, whereas HMGB1 during necrosis may contain all-thiol forms. Moreover, other observations revealed that HMGB1 released during pyroptosis is acetylated. So, HMGB1 released during pyroptosis can subsequently be detected by TLR4 and RAGE receptors to stimulate cytokine release and cell migration.61,62
Emerging roles in the pathogenesis of cancer: HMGB1 as a therapeutic target?
There is growing evidence that HMGB1 may be associated with hallmarks of cancer such as: unlimited replicative potential ability to develop blood vessels (angiogenesis); evasion of programmed cell death (apoptosis); self-sufficiency in growth signals; insensitivity to inhibitors of growth; inflammation; tissue invasion and metastasis; and enhancement of inflammation.63
HMGB1 is one of the most cancer-specific genes. Overexpression of HMGB1 in tumor tissue and increased HMGB1 serum level are near-universal in solid tumors, including colon, gastric, lung, breast, ovarian, pancreatic, and prostate cancer.27
Serum HMGB1 levels were significantly associated with depth of invasion, lymph node metastasis, tumor size, and poor prognosis. The selective upregulation of HMGB1 in tumors contrasts with the other DAMPs, which are expressed under normal conditions and display only modest increased expression in malignancy.64
Interestingly, the mechanisms by which HMGB1 expression is normally suppressed are not fully elucidated. We know that HMGB1 can associate with other molecules, including DNA and cytokines, and activate cells through the differential surface receptors such as TLR4, RAGE, CD24, and TIM-3, influencing the cancer cell’s fate through the cross talk of many different pathways.65
HMGB1 functions as a tumor suppressor in breast cancer, binding to well-known tumor suppressor protein Rb,66 causing Rb-dependent G1 arrest and apoptosis induction. The overexpression of HMGB1 inhibits Rb-positive breast cancer cells growth in vitro, also preventing growth in in vivo tumor models.67
Genomic instability is a hallmark of most cancers.68 Some studies also demonstrate that mammalian HMGB1 and its yeast homologue are critical for maintaining genome stability.68,69 High levels of aneuploidy and spontaneous chromosome aberrations were observed in HMGB1-deficient mouse embryonic fibroblasts (MEFs).69
HMGB1 is involved in major DNA repair pathways, but it could interact with telomerase catalytic component as well as DNA repair enzymes and cofactors to regulate telomere length and DNA repair efficiency.70 The complex roles of HMGB1 in coordinating DNA repair and genomic stability should be further elucidated.
The tumor growth is normally accompanied by reduced microvessel density, therefore resulting in chronic hypoxia. These hypoxic and necrotic regions exhibit increased expression of angiogenic growth factors, recruiting macrophages which produce some angiogenetic cytokines and growth factors.71
Activation of HMGB1 and its receptor RAGE results in the activation of NF-κB pathway, which in turn upregulates leukocyte adhesion molecules and the production of proinflammatory cytokines and angiogenic factors, thereby promoting inflammation and angiogenesis.72 Antibody-targeting HMGB1 inhibits angiogenesis in vitro and in vivo.72
Several HMGB1-targeting agents have been already used in cancer research. These agents include soluble RAGE, HMGB1-neutralizing antibody, platinating agent, ethyl pyruvate, quercetin, and glycyrrhizin.27 HMGB1-neutralizing antibody can block activity of extracellular HMGB1 in tumor therapy.73 Blockade of RAGE signaling pathways could also result in attenuation of tumor development and growth. Several strategies that block HMGB1-RAGE signaling have been described.74 Soluble RAGE acts as a decoy to prevent RAGE signaling and has been used successfully in animal models.75 Platinating agents such as cisplatin and oxaliplatin have the ability to retain HMGB1 within the nucleus.76 HMG domain motifs bind specifically to the major platinum DNA adducts, creating a shield against the human excision nucleases accomplished through the HMGB1 acidic domain.77 Ethyl pyruvate, the first HMGB1 inhibitor used in animal models, inhibits the release of TNF and HMGB1 from endotoxin-stimulated murine macrophages, and also attenuates activation of both the p38 mitogen-activated protein kinase and NF-κB signaling pathways.78 Ethyl pyruvate, interacting with NF-κB, inhibits liver tumor growth.79 In addition, glycyrrhizin and quercetin, potential HMGB1 inhibitors by direct binding to HMGB1 or inhibition of PI3K, improve the effectiveness of anticancer agents in several different tumor models.63 So, targeting the HMGB1 or its receptor represents an important potential application in cancer therapy.80
Conclusion
In 1973, the discovery of HMGB1 as a non-histone chromosomal protein was reported. HMGB1 in the nucleus regulates a number of DNA-related mechanisms.1 In 1999, with the discovery of HMGB1 as a late mediator of sepsis, a new research field was born.28 During the past years, HMGB1 receptors, which balance its activity in multiple cellular processes, have been isolated. In addition to its receptors, HMGB1 posttranslational modifications play an important role in mediating HMGB1 activity.23
Intracellular and extracellular HMGB1 play significantly different roles in inflammation, injury, and cancer. A new function for HMGB1 is as an autophagy regulator, linking the HMGB1 to sterile inflammation, infection, neurodegenerative disease, and cancer.4
Few molecules have as many structures and functions as HMGB1. The understanding of molecular plasticity and multiplicity of HMGB1 is an exciting focus of research, in particular as a prototype for development of anti-inflammatory and immunosuppressive therapies. Future work investigating the details of HMGB1 location, structure, modification, and partners will uncover the secrets of HMGB1’s multiple functions.
Disclosure
The authors report no conflicts of interest in this work.
References
Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973;38(1):14–19. | |
Catena R, Escoffier E, Caron C, Khochbin S, Martianov I, Davidson I. HMGB4, a novel member of the HMGB family, is preferentially expressed in the mouse testis and localizes to the basal pole of elongating spermatids. Biol Reprod. 2009;80(2):358–366. | |
Bustin M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem Sci. 2001;26(3):152–153. | |
Kang R, Chen R, Zhang Q, et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40:1–116. | |
Paonessa G, Frank R, Cortese R. Nucleotide sequence of rat liver HMG1 cDNA. Nucleic Acids Res. 1987;15(21):9077. | |
Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799(1–2):101–113. | |
Bianchi ME, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243(4894 Pt 1):1056–1059. | |
Read CM, Cary PD, Crane-Robinson C, Driscoll PC, Norman DG. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993;21(15):3427–3436. | |
Weir HM, Kraulis PJ, Hill CS, Raine AR, Laue ED, Thomas JO. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993;12(4):1311–1319. | |
Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17(2):72–79. | |
Aizawa S, Nishino H, Saito K, Kimura K, Shirakawa H, Yoshida M. Stimulation of transcription in cultured cells by high mobility group protein 1: essential role of the acidic carboxyl-terminal region. Biochemistry. 1994;33(49):14690–14695. | |
Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19(8):5237–5246. | |
Park JS, Svetkauskaite D, He Q, et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004;279(9):7370–7377. | |
Bierhaus A, Humpert PM, Morcos M, et al. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med (Berl). 2005;83(11):876–886. | |
Yang H, Lundbäck P, Ottosson L, et al. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol Med. 2012;18:250–259. | |
Venereau E, Casalgrandi M, Schiraldi M, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med. 2012;209(9):1519–1528. | |
Bianchi ME. HMGB1 loves company. J Leukoc Biol. 2009;86(3):573–576. | |
Schmidt AM, Yan SD, Yan SF, Stern DM. The biology of the receptor for advanced glycation end products and its ligands. Biochim Biophys Acta. 2000;1498(2–3):99–111. | |
Pedrazzi M, Averna M, Sparatore B, et al. Potentiation of NMDA receptor-dependent cell responses by extracellular high mobility group box 1 protein. PLoS One. 2012;7(8):e44518. | |
Li J, Kokkola R, Tabibzadeh S, et al. Structural basis for the proinflammatory cytokine activity of high mobility group box 1. Mol Med. 2003;9(1–2):37–45. | |
Lu B, Antoine DJ, Kwan K, et al. JAK/STAT1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proc Natl Acad Sci U S A. 2014;111(8):3068–3073. | |
Bustin M, Neihart NK. Antibodies against chromosomal HMG proteins stain the cytoplasm of mammalian cells. Cell. 1979;16(1):181–189. | |
Yang H, Hreggvidsdottir HS, Palmblad K, et al. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc Natl Acad Sci U S A. 2010;107(26):11942–11947. | |
Brezniceanu ML, Völp K, Bösser S, et al. HMGB1 inhibits cell death in yeast and mammalian cells and is abundantly expressed in human breast carcinoma. FASEB J. 2003;17(10):1295–1297. | |
Yuan F, Gu L, Guo S, Wang C, Li GM. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J Biol Chem. 2004;279(20):20935–20940. | |
Travers AA. Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 2003;4(2):131–136. | |
Kang R, Zhang Q, Zeh HJ 3rd, Lotze MT, Tang D. HMGB1 in cancer: good, bad, or both? Clin Cancer Res. 2013;19(15):4046–4057. | |
Wang H, Bloom O, Zhang M, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285(5425):248–251. | |
Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418(6894):191–195. | |
Bell CW, Jiang W, Reich CF 3rd, Pisetsky DS. The extracellular release of HMGB1 during apoptotic cell death. Am J Physiol Cell Physiol. 2006;291(6):C1318–C1325. | |
Kazama H, Ricci JE, Herndon JM, Hoppe G, Green DR, Ferguson TA. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity. 2008;29(1):21–32. | |
Andersson UG, Tracey KJ. HMGB1, a pro-inflammatory cytokine of clinical interest: introduction. J Intern Med. 2004;255(3):318–319. | |
Huang W, Tang Y, Li L. HMGB1, a potent proinflammatory cytokine in sepsis. Cytokine. 2010;51(2):119–126. | |
Passalacqua M, Patrone M, Picotti GB, et al. Stimulated astrocytes release high-mobility group 1 protein, an inducer of LAN-5 neuroblastoma cell differentiation. Neuroscience. 1998;82(4):1021–1028. | |
Faraco G, Fossati S, Bianchi ME, et al. High mobility group box 1 protein is released by neural cells upon different stresses and worsens ischemic neurodegeneration in vitro and in vivo. J Neurochem. 2007;103(2):590–603. | |
Kornblit B, Munthe-Fog L, Madsen HO, Strøm J, Vindeløv L, Garred P. Association of HMGB1 polymorphisms with outcome in patients with systemic inflammatory response syndrome. Crit Care. 2008;12(3):R83. | |
Magna M, Pisetsky DS. The role of HMGB1 in the pathogenesis of inflammatory and autoimmune diseases. Mol Med. 2014;20:138–146. | |
Asavarut P, Zhao H, Gu J, Ma D. The role of HMGB1 in inflammation-mediated organ injury. Acta Anaesthesiol Taiwan. 2013;51(1):28–33. | |
Kohno T, Anzai T, Naito K, et al. Role of high-mobility group box 1 protein in post-infarction healing process and left ventricular remodelling. Cardiovasc Res. 2009;81(3):565–573. | |
Goldstein RS, Gallowitsch-Puerta M, Yang L, et al. Elevated high-mobility group box 1 levels in patients with cerebral and myocardial ischemia. Shock. 2006;25(6):571–574. | |
Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. | |
Ranzato E, Martinotti S, Pedrazzi M, Patrone M. High mobility group box protein-1 in wound repair. Cells. 2012;1(4):699–710. | |
Ranzato E, Patrone M, Pedrazzi M, Burlando B. Hmgb1 promotes wound healing of 3T3 mouse fibroblasts via RAGE-dependent ERK1/2 activation. Cell Biochem Biophys. 2010;57(1):9–17. | |
Ranzato E, Patrone M, Pedrazzi M, Burlando B. HMGb1 promotes scratch wound closure of HaCaT keratinocytes via ERK1/2 activation. Mol Cell Biochem. 2009;332(1–2):199–205. | |
Rovere-Querini P, Capobianco A, Scaffidi P, et al. HMGB1 is an endogenous immune adjuvant released by necrotic cells. EMBO Rep. 2004;5(8):825–830. | |
Bianchi ME, Manfredi AA. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol Rev. 2007;220:35–46. | |
Wittemann B, Neuer G, Michels H, Truckenbrodt H, Bautz FA. Autoantibodies to nonhistone chromosomal proteins HMG-1 and HMG-2 in sera of patients with juvenile rheumatoid arthritis. Arthritis Rheum. 1990;33(9):1378–1383. | |
Kokkola R, Sundberg E, Ulfgren AK, et al. High mobility group box chromosomal protein 1: a novel proinflammatory mediator in synovitis. Arthritis Rheum. 2002;46(10):2598–2603. | |
Pullerits R, Jonsson IM, Verdrengh M, et al. High mobility group box chromosomal protein 1, a DNA binding cytokine, induces arthritis. Arthritis Rheum. 2003;48(6):1693–1700. | |
Popovic K, Ek M, Espinosa A, et al. Increased expression of the novel proinflammatory cytokine high mobility group box chromosomal protein 1 in skin lesions of patients with lupus erythematosus. Arthritis Rheum. 2005;52(11):3639–3645. | |
Andersson A, Covacu R, Sunnemark D, et al. Pivotal advance: HMGB1 expression in active lesions of human and experimental multiple sclerosis. J Leukoc Biol. 2008;84(5):1248–1255. | |
Robinson AP, Caldis MW, Harp CT, Goings GE, Miller SD. High-mobility group box 1 protein (HMGB1) neutralization ameliorates experimental autoimmune encephalomyelitis. J Autoimmun. 2013;43:32–43. | |
Andrassy M, Volz HC, Igwe JC, et al. High-mobility group box-1 in ischemia-reperfusion injury of the heart. Circulation. 2008;117(25):3216–3226. | |
Liesz A, Dalpke A, Mracsko E, et al. DAMP signaling is a key pathway inducing immune modulation after brain injury. J Neurosci. 2015;35(2):583–598. | |
Hoppe G, Talcott KE, Bhattacharya SK, Crabb JW, Sears JE. Molecular basis for the redox control of nuclear transport of the structural chromatin protein Hmgb1. Exp Cell Res. 2006;312(18):3526–3538. | |
Tang D, Kang R, Livesey KM, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190(5):881–892. | |
Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162. | |
Bonaldi T, Talamo F, Scaffidi P, et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22(20):5551–5560. | |
Lu B, Nakamura T, Inouye K, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488(7413):670–674. | |
Antoine DJ, Jenkins RE, Dear JW, et al. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J Hepatol. 2012;56(5):1070–1079. | |
Lamkanfi M, Sarkar A, Vande Walle L, et al. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J Immunol. 2010;185(7):4385–4392. | |
Lamkanfi M. Emerging inflammasome effector mechanisms. Nat Rev Immunol. 2011;11(3):213–220. | |
Tang D, Kang R, Cheh CW, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29(38):5299–5310. | |
Volp K, Brezniceanu ML, Bösser S, et al. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut. 2006;55(2):234–242. | |
Gong H, Zuliani P, Komuravelli A, Faeder JR, Clarke EM. Analysis and verification of the HMGB1 signaling pathway. BMC Bioinformatics. 2010;11 Suppl 7:S10. | |
Jiao Y, Wang HC, Fan SJ. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta Pharmacol Sin. 2007;28(12):1957–1967. | |
Wang LL, Meng QH, Jiao Y, et al. High-mobility group boxes mediate cell proliferation and radiosensitivity via retinoblastoma-interaction-dependent and -independent mechanisms. Cancer Biother Radiopharm. 2012;27(5):329–335. | |
Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability – an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–228. | |
Giavara S, Kosmidou E, Hande MP, et al. Yeast Nhp6A/B and mammalian Hmgb1 facilitate the maintenance of genome stability. Curr Biol. 2005;15(1):68–72. | |
Polanská E, Dobšáková Z, Dvořáčková M, Fajkus J, Štros M. HMGB1 gene knockout in mouse embryonic fibroblasts results in reduced telomerase activity and telomere dysfunction. Chromosoma. 2012;121(4):419–431. | |
Liao D, Johnson RS. Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 2007;26(2):281–290. | |
van Beijnum JR, Buurman WA, Griffioen AW. Convergence and amplification of toll-like receptor (TLR) and receptor for advanced glycation end products (RAGE) signaling pathways via high mobility group B1 (HMGB1). Angiogenesis. 2008;11(1):91–99. | |
Liu L, Yang M, Kang R, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25(1):23–31. | |
Ellerman JE, Brown CK, de Vera M, et al. Masquerader: high mobility group box-1 and cancer. Clin Cancer Res. 2007;13(10):2836–2848. | |
Hanford LE, Enghild JJ, Valnickova Z, et al. Purification and characterization of mouse soluble receptor for advanced glycation end products (sRAGE). J Biol Chem. 2004;279(48):50019–50024. | |
Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399(6737):708–712. | |
Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18(11):1272–1282. | |
Ulloa L, Ochani M, Yang H, et al. Ethyl pyruvate prevents lethality in mice with established lethal sepsis and systemic inflammation. Proc Natl Acad Sci U S A. 2002;99(19):12351–12356. | |
Liang X, Chavez AR, Schapiro NE, et al. Ethyl pyruvate administration inhibits hepatic tumor growth. J Leukoc Biol. 2009;86(3):599–607. | |
Lotze MT, DeMarco RA. Dealing with death: HMGB1 as a novel target for cancer therapy. Curr Opin Investig Drugs. 2003;4(12):1405–1409. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.