Back to Journals » Blood and Lymphatic Cancer: Targets and Therapy » Volume 5
Emerging role of SOX11 in mantle cell lymphoma
Authors Kuci V, Nordstrom L, Jerkeman M, Ek S
Received 10 October 2014
Accepted for publication 21 November 2014
Published 27 February 2015 Volume 2015:5 Pages 35—42
DOI https://doi.org/10.2147/BLCTT.S60447
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor David Dingli
Venera Kuci,1,2,* Lena Nordström,1,2,* Mats Jerkeman,3 Sara Ek1,2
1Department of Immunotechnology, Lund University, Lund, Sweden; 2CREATE Health, Lund University, Lund, Sweden; 3Department of Oncology, Lund University, Lund, Sweden
*These authors contributed equally to this work
Abstract: During recent years the neural transcription factor SOX11 has been established as an important biomarker for mantle cell lymphoma. SOX11 is both a diagnostic and prognostic antigen, and may potentially be used for treatment selection for younger patients, in relation to protocols including high dose chemotherapy. The molecular pathways involved are still not fully elucidated and, as SOX11 can interact with several co-transcription factors, functional assays need to be carefully designed to pinpoint SOX11-specific function in a defined cellular context. Furthermore, as SOX11 belongs to a large family of homologous proteins, analysis of SOX11 has been limited by the availability of specific antibodies for detection and pull-down. In this review, we discuss the emerging role of SOX11 in mantle cell lymphoma and discuss the potential impact in relation to tumorigenesis, diagnostics, prognostics, and therapy.
Keywords: SOX11, mantle cell lymphoma, diagnostic
Mantle cell lymphoma – is B cell receptor function and CCND1 overexpression necessary for tumorigenesis?
Mantle cell lymphoma (MCL) is a very aggressive B-cell lymphoma with patients having a median age of 68 years.1 Previously, MCL has been considered an incurable disease, with a median survival slowly increasing from 3 years in the 1980s to around 5 years in the early 2000s.2 However, recent new treatment protocols combining high dose cytarabine with autologous stem cell transplantation and rituximab has achieved significant improvement in long-term survival in younger patients. One such study, the Nordic MCL2 trial, has shown very encouraging results with a median survival of more than 10 years.3
MCL was originally considered to be derived from naïve B cells and most tumors show unmutated immunoglobulin (Ig) VH genes. However, later studies have shown that 10%−20% of MCLs have mutated Ig genes, indicating that a sub-group has passed through a differentiation stage involving somatic hypermutation of VH genes.4–9 It is noteworthy that all MCL cells in an individual, including the ones that are VH mutated, are clonally identical, indicating that the tumor cells are frozen at the stage where the malignant transformation took place.10,11 Interestingly, it has been reported that B cells can mutate their VH genes in the absence of GC, but the mechanisms involved are far from understood.12,13 Furthermore, a biased VH usage has been described for MCL, which suggests that the transformation is antigen driven,5,7,8,11 as recently discussed by Hadzidimitriou et al.14 No antigen has been identified but VH 3-21, which is one of the most commonly used VH genes in MCL, has been used to produce rheumatoid factors and may recognize an auto-antigen.10,15 Most lymphomas express a functional B cell receptor due to successful rearrangement of one allele, despite the occurrence of translocations involving the Ig heavy chain locus on the other allele, indicating that, at least initially, the B cell was dependent on a functional B cell receptor.16 This is further supported by the successful use of BTK inhibitors, such as ibrutinib, in treatment of MCL.17
Cyclin D1 is the hallmark of MCL, and the overexpression is most often caused by a translocation of the CCND1 gene to the IgH (11;14). The translocation occurs during an attempted primary DH-JH rearrangement in early B cells, but may also occur later during a secondary rearrangement.4 Although the majority of MCL patients show overexpression of Cyclin D1, there are Cyclin D1 negative MCL-like cases that have an indolent course of disease.18,19 For some Cyclin D1 negative cases, it has been shown that they have similar clinicopathological and gene expression profiles as conventional Cyclin D1 positive MCL but overexpressing Cyclin D2 and Cyclin D3, supporting a role for the D-type cyclins in the pathogenesis of MCL.20
However, although Cyclin D1 is overexpressed in MCL, it has been shown that Cyclin D1 does not induce MCL development in vivo.21 Furthermore, the lack of prognostic information related to Cyclin D122 and the existence of Cyclin D1 negative MCL (7%–15%) challenges the requisite of this protein for tumorigenesis.18,23 Knockdown of Cyclin D1 has also been reported to only have a limited effect on cell proliferation and apoptosis.24 Furthermore, healthy individuals may carry B-cells with the t(11;14) translocations, supporting the need for additional genetic hits for lymphomagenesis.25
It is possible that such additional oncogenic events may precede the translocation of CCND1 and allow it to occur unhindered. One mechanism for this could be the frequent mutation/deletion of ataxia telangiectasia mutated (ATM).26,27 ATM, which is involved in the cellular responses to double-strand breaks, is normally expressed in pre-GC B cells and it has been suggested that inactivation of the gene may be involved in malignant transformation.28,29
In MCL, it has been assumed that Cyclin D1 primarily acts through its role in the cell cycle, controlling the transition of G1 to S phase by binding and activating cyclin-dependent kinase (CDK) 4 and CDK6 followed by phosphorylation of the retinoblastoma protein and release of the transcription factor E2F.30 However, in other malignancies several CDK-independent functions of Cyclin D1 have been identified, including interaction with the pro-apoptotic protein BAX21 and RAD51,31 the latter impacting DNA repair. This potential involvement of Cyclin D1 in apoptosis and/or DNA repair pathways in MCL is highly interesting but unexplored.
SOX11 and its role as a routine diagnostic tool in MCL
The neural transcription factor SOX11 has during recent years been shown to be aberrantly expressed in several types of cancers, such as medulloblastoma,32 glioma,33 MCL,34 and epithelial ovarian cancer,35,36 while corresponding non-malignant tissues lack expression.
The Sox family belongs to a group of 20 TFs within the high-mobility group (HMG) box protein super family, which are characterized by high sequence homology within their DNA-binding HMG domain.37 It has been proposed that the HMG domain serves two functions, ie, DNA binding and partner selection, which may permit selective recruitment of SOX proteins to specific promoters and transcription factors. To date, the main known function of SOX11 has been its importance for neural development38 and organogenesis39 during fetal development. Recent data also suggest an important role for SOX11 as a transcriptional regulator in adult immature neurons.40 Of note, limited information on the role of SOX11 in normal and malignant tissues are available and present data point towards a dual role in relation to survival and growth consistent with previous reports of SOX proteins being promiscuous in the DNA binding,41 allowing use of tissue-specific co-transcription factors. So far, functional investigations of SOX11 in both similar and separate cellular contexts show diverse results. This may be explained by the potential use of different co-transcription factors but also by technical challenges to specifically pin-point SOX11 among other homologous SOX members.
In MCL, SOX11 is a disease-associated antigen and can be used to diagnose this subtype with high accuracy using immunohistochemistry,34,42 as confirmed by multiple independent laboratories.43–45 SOX11 has shown to be an important tool to accurately diagnose the small fraction of Cyclin D1 negative MCL cases.42,44 It has also been proposed to correlate to unmutated disease and distinguish rare indolent SOX11 negative MCL from classical MCL,46,47 which may have an impact on treatment selection. Subsequently, novel methods and improved tools have been developed to be able to clinically use this biomarker for diagnostic and prognostic purposes. Among others, qPCR-based detection of SOX1148 and methods to analyze fine needle aspirates49 have been developed. It has further been suggested that SOX11 may be used as a marker for minimal residual disease.50,51
Analysis of SOX11 protein, being highly homologous to other SOX4 and SOX12, has been hampered by unspecific tools and only recently a specific monoclonal antibody was developed.50 The superiority of this antibody has also been confirmed by independent users.52 Thus, current literature should be carefully interpreted to acknowledge that analysis may include background levels of other SOX family proteins.
In summary, SOX11 has been identified as an important MCL-associated antigen that complements Cyclin D1 overexpression to accurately diagnose MCL and to distinguish it from molecular mimics such as CLL. In unselected cohorts, the numbers of SOX11 negative cases are low and it has been proposed that these might be transformed cases from indolent SOX11 negative disease. If that holds true, SOX11 might be regarded as a stand-alone diagnostic antigen for de-novo classical MCL.
SOX11 as a prognostic tool
The portfolio of MCL treatment strategies has increased dramatically during the last decade and includes high dose chemotherapy, stem cell transplantation, immunomodulatory agents, as well as targeted therapies such as rituximab, bortezomib, and ibrutinib.17,53 However, there is currently no standard therapy available and treatment selection depends mostly on age and performance status. With a growing arsenal of effective agents – some even providing a possible cure for subgroups of patients – the need for prognostic and predictive biomarkers is urged to guide clinicians in selecting an optimal therapy for each patient.
The MCL International Prognostic Index (MIPI) was developed in 2008 and is based on the independent prognostic factors, age, performance status (ECOG PS), lactate dehydrogenase and white blood cell count.54 MIPI categorizes patients into low, intermediate, and high risk groups and its prognostic value was recently confirmed in a large set of MCL patients included in the European MCL Network.55 Although MIPI is a part of the routine work-up in most clinics, it is still rarely used for treatment decisions.56 Therefore, increased efforts in identifying biomarkers useful as companion diagnostics are needed. In this case, biological biomarkers, which reflect the underlying pathogenesis of MCL, have a huge potential.
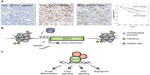
The prognostic impact of SOX11 has been controversial, being associated with both an improved and inferior survival. Initial studies have assessed survival in population-based cohorts, including both indolent and aggressive MCL. SOX11 has been associated with improved overall survival in two studies including 53 and 186 patients respectively.45,57 In contrast, SOX11 expression has also been identified as a discriminating antigen between indolent MCL (SOX11 negative) and aggressive MCL (SOX11 positive), thus correlating SOX11 expression to poorer outcome and lack of hypermutation of VH genes.46,47 This is in contrary to studies presented by Nygren et al and Kuo et al, which concluded that most of their indolent MCL patients were SOX11 positive.49,57 Recently, two independent studies associated increased SOX11 expression with improved overall survival in patients treated with either the Nordic protocol22 (Figure 1A) or with R-Hyper-CVAD49 pin-pointing a role of SOX11 as a predictive biomarker for these treatments.
To reach consensus on the prognostic relevance of SOX11, several international guidelines need to be established. Indolent MCLs display a large patient heterogeneity within different studies and international guidelines defining indolent MCL need to be improved and well-defined patient cohorts, treated with modern protocols, use to further investigate the prognostic relevance of SOX11. Furthermore, as most studies use IHC to assess SOX11 expression, it is worth mentioning that tissue fixation time could have a large impact on the staining intensity. Also, most studies have assessed SOX11 expression using polyclonal reagents, which display a potential cross-reactivity to SOX450 and/or suffer from batch variations. Thus, current literature supports the use of specific monoclonal SOX11 antibodies that today are present on the market.22,52
Besides SOX11, several other molecular biomarkers have been suggested to add prognostic information in relation to MCL. Ki-67 has previously shown prognostic value, and when combined with MIPI, form the biological MIPI.54
Another protein commonly affected in tumorigenesis is p53 and disruption of the p53 pathway is a common event in MCL. TP53 is one of the most frequently mutated genes (14%–20%) in MCL patients58–60 and both TP53 mutations and strong expression of the protein have been correlated to shorter survival22,59 and high proliferation.61 In our recent study, we explored the impact of the molecular markers Ki-67, Cyclin D1, SOX11, and p53 to add prognostic power to MIPI using multivariate analysis.22 P53 was the only molecule that remained significant and the combined p53-MIPI was able to identify a low risk group with longer overall survival and a high risk group with significantly shorter event-free survival in patients treated according to the Nordic protocol.
To fully explore the potential of SOX11, p53, Ki-67, and other novel biomarkers, well defined prospective studies needs to be conducted and validated by independent research groups.
Epigenetic regulation of SOX11
Transcription factors and chromatin modifying enzymes drive cellular identity, dictating either activation or repression of specific transcriptional programs. The aberrant expression of SOX11 is not a direct consequence of the genetic instability in MCL and no copy number alterations or mutations have been reported. In chicken germ cells, SOX11 has been shown to be regulated by miR-302a and miR-456,62 but it is unknown if this is species and/or tissue specific as these particular micro ribonucleic acids (miRNAs) were not identified by Navarro et al comparing microRNA expression among SOX11 positive and negative MCL tumors.63 Initial investigations on SOX11 promoter methylation in B-cell lymphomas revealed a distinct difference in promoter methylation where SOX11 expressing MCLs have an unmethylated promoter, and non-expressing lymphomas have a strongly methylated promoter.64 Vegliante et al further demonstrated that the rare subgroup of SOX11 negative MCLs as well as normal peripheral B-cells display an unmethylated promoter and that transcription was silenced by repressive histone marks (as schematically illustrated in Figure 1B).65 Interestingly, they further showed that SOX11 expression is induced in hematopoietic cells by expression of OCT4, KLF4, SOX2, and c-MYC in induced pluripotent cells.65 However, OCT4, KLF4, and SOX2 are not expressed in MCL cell-lines.66
Although SOX11 is silenced in most adult tissue, the promoter remains unmethylated (unpublished data, 2014). However, many tumors methylate SOX11 de novo during their pathogenesis as reported in B-cell lymphomas (excluding MCL)64 and subgroups of epithelial ovarian cancer.36 Additionally, SOX11 methylation has been correlated to lymph node metastasis in nasopharyngeal carcinoma67 and included in a five gene signature for detection of bladder cancer.68
With the growing interest in using epigenetic therapies in hematological malignancies, investigations on genetic and epigenetic regulation of SOX11 are needed to understand its aberrant expression in disease. While the epigenetic regulation of SOX11 has been initially described, no studies have investigated SOX11 upstream regulatory transcription factors in MCL.
The molecular function of SOX11 in MCL
Improved treatment strategies for MCL patients could be developed if the specific molecular drivers of disease were identified. Among others, novel therapies targeting BTK (ibrutinib) and BCL-2 (ABT-199) show promising effects and may in the future contribute to improved treatment success for MCL patients.
The molecular function of SOX11 has so far mainly been investigated in the normal setting, and proved to be involved in adult and embryogenic neurogenesis.69 However, as SOX11 is promiscuous in its DNA binding capacity,70 it is expected that down-stream targets are tissue specific. Thus, functional studies of SOX11 need to be performed in MCL to identify involved signaling pathways. In such recent efforts, several important molecular mechanisms of SOX11 have been revealed. Several independent studies have used gene expression profiling (GEP) on SOX11 silenced and overexpressed models to identify pathways involved in SOX11 regulation. Of interest, the Cyclin D1 related pathway Rb-E2F was shown to be affected by the increased level of SOX11 upon transient overexpression.64 Biologically similar results were shown using a knock-down model where a number of genes, among others DBN1, previously associated with MCL and involved in migration, proliferation, and apoptosis was co-regulated with SOX11,71 pinpointing the global impact of SOX11 on MCL-associated gene expression. In this stable knock-down model, it was further shown that SOX11 reduction caused increased growth, partially driven by the upregulation of autotaxin (ATX).71 Of interest, ATX has previously has been reported to be involved in tumor aggressiveness and angiogenesis.72
Besides GEP, chromatin immunoprecipitation (ChIP) has been used alone, or in combination with GEP to assess the transcription factor binding sites of SOX11. Using these technologies, DBN1, SETMAR, and HIG2 were identified as direct targets of SOX11.73 Further studies using ChIP-seq has pin-pointed WNT signaling as a potential important pathway down-stream of SOX11.49 In a separate study by Vegliante et al, they suggest that SOX11 blocks terminal B-cell differentiation in aggressive MCL through direct positive regulation of PAX5.74 It is known that SOX genes are promiscuous in their DNA binding capacity and thus, it is not surprising that several pathways may be affected – but it remains to be determined which ones are of most biological value for MCL pathogenesis (see schematic illustration in Figure 1C). It should be stressed that i) MCL cell lines are molecularly different in various aspects and ii) that immunoprecipitation assays are highly dependent on the specificity of the used antibody for enrichment. Thus, the use of single or few cell lines and the potential use of different and cross-reactive antibodies may explain the poor overlap between GEP and ChIP studies.
Recent in vivo studies of SOX11 have revealed a role in angiogenesis, where SOX11-positive MCL cells promote vasculature development through regulation of PDGFA.75 Furthermore, in that experimental set-up SOX11 is associated with increased growth,74 in contrast to previous observations where SOX11 is associated with decreased growth, lower tumorigenicity, and longer time to symptoms of disease compared to SOX11 silenced MCL cells.71 The development of efficient anti-angiogenic treatment is warranted, but so far the clinical benefit of anti-angiogenic treatment in MCL has been limited,76 although thalidomide has shown some effect in relapsed patients.77
Conclusion
SOX11 has emerged as a highly interesting disease-defining marker for MCL and detailed investigations of involved signaling pathways are warranted to identify potential targets for improved intervention. It is evident that we need to look beyond the apparent conflict between SOX11 as i) a disease-defining antigen for classical MCL and ii) associated with improved prognosis within this diagnostic group. Efforts to identify a molecular link between p53, Cyclin D1, and/or SOX11 have so far not been successful but need to be carefully investigated to, if possible, identify synthetically lethal combinations for MCL cells that can lead to a cure.
Disclosure
A patent has previously been filed on the use of SOX11 as a diagnostic, prognostic, and therapeutic target in MCL. The authors report no other conflict of interest in this work.
References
Caballero D, Campo E, Lopez-Guillermo A, et al. Clinical practice guidelines for diagnosis, treatment, and follow-up of patients with mantle cell lymphoma. Recommendations from the GEL/TAMO Spanish Cooperative Group. Ann Hematol. 2013;92(9):1151–1179. | |
Herrmann A, Hoster E, Zwingers T, et al. Improvement of overall survival in advanced stage mantle cell lymphoma. J Clin Oncol. 2009;27(4):511–518. | |
Geisler CH, Kolstad A, Laurell A, et al. Nordic MCL2 trial update: six-year follow-up after intensive immunochemotherapy for untreated mantle cell lymphoma followed by BEAM or BEAC + autologous stem-cell support: still very long survival but late relapses do occur. Br J Haematol. 2012;158(3):355–362. | |
Welzel N, Le T, Marculescu R, et al. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001;61(4):1629–1636. | |
Pittaluga S, Tierens A, Pinyol M, Campo E, Delabie J, De Wolf-Peeters C. Blastic variant of mantle cell lymphoma shows a heterogenous pattern of somatic mutations of the rearranged immunoglobulin heavy chain variable genes. Br J Haematol. 1998;102(5):1301–1306. | |
Laszlo T, Nagy M, Kelenyi G, Matolcsy A. Immunoglobulin V(H) gene mutational analysis suggests that blastic variant of mantle cell lymphoma derives from different stages of B-cell maturation. Leuk Res. 2000;24(1):27–31. | |
Thorselius M, Walsh S, Eriksson I, et al. Somatic hypermutation and V(H) gene usage in mantle cell lymphoma. Eur J Haematol. 2002;68(4):217–224. | |
Camacho FI, Algara P, Rodriguez A, et al. Molecular heterogeneity in MCL, defined by the use of specific VH genes and the frequency of somatic mutations. Blood. Epub January 2, 2003. | |
Kienle D, Krober A, Katzenberger T, et al. VH mutation status and VDJ rearrangement structure in mantle cell lymphoma: correlation with genomic aberrations, clinical characteristics, and outcome. Blood. 2003;102(8):3003–3009. | |
Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2(12):920–932. | |
Walsh SH, Thorselius M, Johnson A, et al. Mutated VH genes and preferential VH3-21 use define new subsets of mantle cell lymphoma. Blood. 2003;101(10):4047–4054. | |
Weller S, Faili A, Garcia C, et al. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci U S A. 2001;98(3):1166–1170. | |
Matsumoto M, Lo SF, Carruthers CJ, et al. Affinity maturation without germinal centres in lymphotoxin-alpha-deficient mice. Nature. 1996;382(6590):462–466. | |
Hadzidimitriou A, Agathangelidis A, Darzentas N, et al. Is there a role for antigen selection in mantle cell lymphoma? Immunogenetic support from a series of 807 cases. Blood. 2011;118(11):3088–3095. | |
He X, Goronzy JJ, Zhong W, Xie C, Weyand CM. VH3-21 B cells escape from a state of tolerance in rheumatoid arthritis and secrete rheumatoid factor. Mol Med. 1995;1(7):768–780. | |
Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90(6):1073–1083. | |
Shah N, Hutchinson C, Rule S. Ibrutinib for the treatment of mantle cell lymphoma. Expert Rev Hematol. 2014;7(5):521–531. | |
Yatabe Y, Suzuki R, Tobinai K, et al. Significance of cyclin D1 overexpression for the diagnosis of mantle cell lymphoma: a clinicopathologic comparison of cyclin D1-positive MCL and cyclin D1-negative MCL-like B-cell lymphoma. Blood. 2000;95(7):2253–2261. | |
Hashimoto Y, Nakamura N, Kuze T, Abe M. The evaluation of the biological behavior and grade among cases with mantle cell lymphoma. Leuk Lymphoma. 2002;43(3):523–530. | |
Fu K, Weisenburger DD, Greiner TC, et al. Cyclin D1-negative mantle cell lymphoma: a clinicopathologic study based on gene expression profiling. Blood. 2005;106(13):4315–4321. | |
Beltran E, Fresquet V, Martinez-Useros J, et al. A cyclin-D1 interaction with BAX underlies its oncogenic role and potential as a therapeutic target in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2011;108(30):12461–12466. | |
Nordstrom L, Sernbo S, Eden P, et al. SOX11 and TP53 add prognostic information to MIPI in a homogenously treated cohort of mantle cell lymphoma – a Nordic Lymphoma Group study. Br J Haematol. 2014;166(1):98–108. | |
Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–197. | |
Klier M, Anastasov N, Hermann A, et al. Specific lentiviral shRNA-mediated knockdown of cyclin D1 in mantle cell lymphoma has minimal effects on cell survival and reveals a regulatory circuit with cyclin D2. Leukemia. 2008;22(11):2097–2105. | |
Lecluse Y, Lebailly P, Roulland S, Gac AC, Nadel B, Gauduchon P. t(11;14)-positive clones can persist over a long period of time in the peripheral blood of healthy individuals. Leukemia. 2009;23(6):1190–1193. | |
Schaffner C, Idler I, Stilgenbauer S, Dohner H, Lichter P. Mantle cell lymphoma is characterized by inactivation of the ATM gene. Proc Natl Acad Sci U S A. 2000;97(6):2773–2778. | |
Fang NY, Greiner TC, Weisenburger DD, et al. Oligonucleotide microarrays demonstrate the highest frequency of ATM mutations in the mantle cell subtype of lymphoma. Proc Natl Acad Sci U S A. 2003;100(9):5372–5377. | |
Starczynski J, Simmons W, Flavell JR, et al. Variations in ATM protein expression during normal lymphoid differentiation and among B-cell-derived neoplasias. Am J Pathol. 2003;163(2):423–432. | |
Bertoni F, Zucca E, Cotter FE. Molecular basis of mantle cell lymphoma. Br J Haematol. 2004;124(2):130–140. | |
Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15(2):158–163. | |
Jirawatnotai S, Hu Y, Michowski W, et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474(7350):230–234. | |
Lee CJ, Appleby VJ, Orme AT, Chan WI, Scotting PJ. Differential expression of SOX4 and SOX11 in medulloblastoma. J Neurooncol. 2002;57(3):201–214. | |
Weigle B, Ebner R, Temme A, et al. Highly specific overexpression of the transcription factor SOX11 in human malignant gliomas. Oncol Rep. 2005;13(1):139–144. | |
Ek S, Dictor M, Jerkeman M, Jirstrom K, Borrebaeck CA. Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood. 2008;111(2):800–805. | |
Brennan DJ, Ek S, Doyle E, et al. The transcription factor Sox11 is a prognostic factor for improved recurrence-free survival in epithelial ovarian cancer. Eur J Cancer. 2009;45(8):1510–1517. | |
Sernbo S, Gustavsson E, Brennan DJ, et al. The tumour suppressor SOX11 is associated with improved survival among high grade epithelial ovarian cancers and is regulated by reversible promoter methylation. BMC Cancer. 2011;11:405. | |
Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27(6):1409–1420. | |
Azuma T, Ao S, Saito Y, et al. Human SOX11, an upregulated gene during the neural differentiation, has a long 3′ untranslated region. DNA Res. 1999;6(5):357–360. | |
Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and SOX proteins in glial cells. J Biol Chem. 1998;273(26):16050–16057. | |
Haslinger A, Schwarz TJ, Covic M, Lie DC. Expression of Sox11 in adult neurogenic niches suggests a stage-specific role in adult neurogenesis. Eur J Neurosci. 2009;29(11):2103–2114. | |
Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39(12):2195–2214. | |
Dictor M, Ek S, Sundberg M, et al. Strong lymphoid nuclear expression of SOX11 transcription factor defines lymphoblastic neoplasms, mantle cell lymphoma and Burkitt’s lymphoma. Haematologica. 2009;94(11):1563–1568. | |
Chen YH, Gao J, Fan G, Peterson LC. Nuclear expression of sox11 is highly associated with mantle cell lymphoma but is independent of t(11;14)(q13;q32) in non-mantle cell B-cell neoplasms. Mod Pathol. 2010;23(1):105–112. | |
Mozos A, Royo C, Hartmann E, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica. 2009;94(11):1555–1562. | |
Wang X, Asplund AC, Porwit A, et al. The subcellular Sox11 distribution pattern identifies subsets of mantle cell lymphoma: correlation to overall survival. Br J Haematol. 2008;143(2):248–252. | |
Fernandez V, Salamero O, Espinet B, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70(4):1408–1418. | |
Navarro A, Clot G, Royo C, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res. 2012;72(20):5307–5316. | |
Hamborg KH, Bentzen HH, Grubach L, Hokland P, Nyvold CG. A highly sensitive and specific qPCR assay for quantification of the biomarker SOX11 in mantle cell lymphoma. Eur J Haematol. 2012;89(5):385–394. | |
Kuo PY, Leshchenko VV, Fazzari MJ, et al. High-resolution chromatin immunoprecipitation (ChIP) sequencing reveals novel binding targets and prognostic role for SOX11 in mantle cell lymphoma. Oncogene. Epub March 31, 2014. | |
Nordstrom L, Andreasson U, Jerkeman M, Dictor M, Borrebaeck C, Ek S. Expanded clinical and experimental use of SOX11 – using a monoclonal antibody. BMC Cancer. 2012;12:269. | |
Simonsen AT, Sorensen CD, Ebbesen LH, Bodker JS, Bentzen HH, Nyvold CG. SOX11 as a minimal residual disease marker for Mantle cell lymphoma. Leuk Res. 2014;38(8):918–924. | |
Nakashima MO, Durkin L, Bodo J, et al. Utility and Diagnostic Pitfalls of SOX11 Monoclonal Antibodies in Mantle Cell Lymphoma and Other Lymphoproliferative Disorders. Appl Immunohistochem Mol Morphol. 2014;22(10):720–727. | |
Moshaver B, van Rhenen A, Kelder A, et al. Identification of a small subpopulation of candidate leukemia-initiating cells in the side population of patients with acute myeloid leukemia. Stem Cells. 2008;26(12):3059–3067. | |
Hoster E, Dreyling M, Klapper W, et al. A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood. 2008;111(2):558–565. | |
Hoster E, Klapper W, Hermine O, et al. Confirmation of the mantle-cell lymphoma International Prognostic Index in randomized trials of the European Mantle-Cell Lymphoma Network. J Clin Oncol. 2014;32(13):1338–1346. | |
Dreyling M, Kluin-Nelemans HC, Bea S, et al. Update on the molecular pathogenesis and clinical treatment of mantle cell lymphoma: report of the 11th annual conference of the European Mantle Cell Lymphoma Network. Leuk Lymphoma. 2013;54(4):699–707. | |
Nygren L, Baumgartner Wennerholm S, Klimkowska M, Christensson B, Kimby E, Sander B. Prognostic role of SOX11 in a population-based cohort of mantle cell lymphoma. Blood. 2012;119(18):4215–4223. | |
Greiner TC, Dasgupta C, Ho VV, et al. Mutation and genomic deletion status of ataxia telangiectasia mutated (ATM) and p53 confer specific gene expression profiles in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2006;103(7):2352–2357. | |
Halldorsdottir AM, Lundin A, Murray F, et al. Impact of TP53 mutation and 17p deletion in mantle cell lymphoma. Leukemia. 2011; 25(12):1904–1908. | |
Zhang J, Jima D, Moffitt AB, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123(19):2988–2996. | |
Slotta-Huspenina J, Koch I, de Leval L, et al. The impact of cyclin D1 mRNA isoforms, morphology and p53 in mantle cell lymphoma: p53 alterations and blastoid morphology are strong predictors of a high proliferation index. Haematologica. 2012;97(9):1422–1430. | |
Lee SI, Lee BR, Hwang YS, et al. MicroRNA-mediated posttranscriptional regulation is required for maintaining undifferentiated properties of blastoderm and primordial germ cells in chickens. Proc Natl Acad Sci U S A. 2011;108(26):10426–10431. | |
Navarro A, Clot G, Prieto M, et al. microRNA expression profiles identify subtypes of mantle cell lymphoma with different clinicobiological characteristics. Clin Cancer Res. 2013;19(12):3121–3129. | |
Gustavsson E, Sernbo S, Andersson E, et al. SOX11 expression correlates to promoter methylation and regulates tumor growth in hematopoietic malignancies. Mol Cancer. 2010;9:187. | |
Vegliante MC, Royo C, Palomero J, et al. Epigenetic Activation of SOX11 in Lymphoid Neoplasms by Histone Modifications. PLoS One. 2011;6(6):e21382. | |
Rahal R, Frick M, Romero R, et al. Pharmacological and genomic profiling identifies NF-kappaB-targeted treatment strategies for mantle cell lymphoma. Nat Med. 2014;20(1):87–92. | |
Zhang S, Li S, Gao JL. Promoter methylation status of the tumor suppressor gene SOX11 is associated with cell growth and invasion in nasopharyngeal carcinoma. Cancer Cell Int. 2013;13(1):109. | |
Chung W, Bondaruk J, Jelinek J, et al. Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol Biomarkers Prev. 2011;20(7):1483–1491. | |
Wang Y, Lin L, Lai H, Parada LF, Lei L. Transcription factor Sox11 is essential for both embryonic and adult neurogenesis. Dev Dyn. 2013;242(6):638–653. | |
Wiebe MS, Nowling TK, Rizzino A. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. J Biol Chem. 2003;278(20):17901–17911. | |
Conrotto P, Andreasson U, Kuci V, Borrebaeck CA, Ek S. Knock-down of SOX11 induces autotaxin-dependent increase in proliferation in vitro and more aggressive tumors in vivo. Mol Oncol. 2011;5(6):527–537. | |
Nam SW, Clair T, Kim YS, et al. Autotaxin (NPP-2), a metastasis-enhancing motogen, is an angiogenic factor. Cancer Res. 2001;61(18):6938–6944. | |
Wang X, Bjorklund S, Wasik AM, et al. Gene expression profiling and chromatin immunoprecipitation identify DBN1, SETMAR and HIG2 as direct targets of SOX11 in mantle cell lymphoma. PLoS One. 2010;5(11):e14085. | |
Vegliante MC, Palomero J, Perez-Galan P, et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood. 2013;121(12):2175–2185. | |
Palomero J, Vegliante MC, Rodriguez ML, et al. SOX11 promotes tumor angiogenesis through transcriptional regulation of PDGFA in mantle cell lymphoma. Blood. 2014;124(14):2235–2247. | |
Ribatti D, Nico B, Ranieri G, Specchia G, Vacca A. The role of angiogenesis in human non-Hodgkin lymphomas. Neoplasia. 2013;15(3):231–238. | |
Kaufmann H, Raderer M, Wohrer S, et al. Antitumor activity of rituximab plus thalidomide in patients with relapsed/refractory mantle cell lymphoma. Blood. 2004;104(8):2269–2271. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.