Back to Journals » Infection and Drug Resistance » Volume 12
Emergence of two Escherichia coli strains co-harboring mcr-1 and blaNDM in fresh vegetables from China
Received 11 April 2019
Accepted for publication 30 July 2019
Published 23 August 2019 Volume 2019:12 Pages 2627—2635
DOI https://doi.org/10.2147/IDR.S211746
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Bao-Tao Liu,1 Feng-Jing Song2
1Department of Basic Veterinary Medicine, College of Veterinary Medicine, Qingdao Agricultural University, Qingdao, People’s Republic of China; 2Institute of Plant Protection, Qingdao Academy of Agricultural Sciences, Qingdao, People’s Republic of China
Correspondence: Bao-Tao Liu
Department of Basic Veterinary Medicine, College of Veterinary Medicine, Qingdao Agricultural University, Qingdao 266109, People’s Republic of China
Tel +86 1 346 587 7602
Email [email protected]
Feng-Jing Song
Institute of Plant Protection, Qingdao Academy of Agricultural Sciences, Qingdao, People’s Republic of China
Email [email protected]
Background: The concurrence of mcr and carbapenemase genes among Enterobacteriaceae has been a great clinical concern. In our study, we aimed to investigate the prevalence of mcr-positive carbapenem-resistant Enterobacteriaceae (CRE) in fresh vegetables and shed light on the possibility of transmission of mcr-positive CRE via fresh vegetables.
Methods: In this study, 712 fresh vegetable samples from 10 provinces in China were collected between May 2017 and Dec 2018 and were screened for mcr and carbapenemase genes. Antibiotic susceptibilities for isolates co-harboring carbapenemase genes and mcr were determined by an agar dilution or a broth microdilution method. Pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) analysis were also performed. Transferability of the carbapenemase/mcr-bearing plasmids was determined by conjugation, replicon typing and S1-PFGE-Southern blotting. The sequences of these plasmids were analyzed by using whole-genome sequencing with Illumina Hiseq platform.
Results: Two E. coli isolates concomitantly carrying mcr-1 and blaNDM-5/9 from leaf rape and spinach, respectively, were found and both isolates showed multidrug resistance. Notably, mcr-1-positive 690 harboring blaNDM-5 and 701 carrying blaNDM-9 belonged to ST156 and ST2847, respectively, similar to the prevalent MLST types of E. coli co-carrying mcr-1 and blaNDM from avian in our previous study. mcr-1 was on ∼33-kb IncX4 plasmid or ∼60-kb IncI2 plasmid, while blaNDM-5/9 was on ∼46-kb IncX3 plasmid or ∼120-kb untypable plasmid. The plasmids were highly similar to those from animals and clinical patients reported in various countries.
Conclusion: E. coli isolates concomitantly carrying mcr-1 and blaNDM-5/9 in fresh vegetables may serve as a direct source of pathogens in humans, and such discovery in fresh vegetables emphasizes the importance of prompt surveillance and intervention in limiting the spread of E. coli co-carrying blaNDM and mcr-1. To our knowledge, this is the first report of Enterobacteriaceae co-carrying blaNDM and mcr-1 in fresh vegetables.
Keywords: carbapenem resistance, colistin resistance, coexistence, plasmids, Enterobacteriaceae
Introduction
With the increasing carbapenem consumption in the past two decades, unprecedented global increase has been observed in the populations of carbapenem-resistant Enterobacteriaceae (CRE), posing colistin as the last therapeutic resort for the treatment of such organism. However, the efficacy of the drug has been challenged by the emergence of mobilized colistin resistance (mcr) genes.1 Of great clinical concern is the concurrence of mcr and carbapenemase genes among Enterobacteriaceae. In fact, mcr has been found in CRE isolates from food animals2 and humans,3–6 around the world, especially in China. High prevalence of mcr-positive CRE isolates has been found among various origins in China, including humans,3,7–9 retail meat,10 food animals,11,12 dogs,11 birds and flies.11
Consumption of fresh vegetables has increased over the recent years, because vegetables can provide essential components for humans.13 However, fresh vegetables eaten raw have been linked with outbreak of foodborne diseases14 and have often served as resistance gene “reservoirs”. For example, cephalosporin-resistant Enterobacteriaceae were found on 5.2% of the 1216 vegetables obtained from Dutch stores during 2012 and 2013.15 About 25.4% of the 169 vegetables imported from the Dominican Republic, India, Thailand and Vietnam in 2014 harbored one or more extended-spectrum-beta-lactamase-producing Enterobacteriaceae.16
Recently, mcr-positive isolates have been also found in one lettuce sample in Portugal17 and in two imported vegetable samples in Switzerland.18 In China, 9 of the 916 vegetables (0.98%) sampled in Guangzhou carried mcr-1,19 while 19 of the 528 fresh vegetables (3.60%) were found to harbor mcr-1-positive isolates in our previous report.20 In addition, CRE isolates were also sporadically found in fresh vegetables, including one blaOXA-181-positive Klebsiella variicola in Switzerland,21 three blaOXA-181-positive K. pneumoniae in Algeria,22 one E. coli co-harboring blaNDM-1 and blaKPC-2 in China23 and twelve isolates in our previous study in China.24 All these findings suggest that Enterobacteriaceae-producing carbapenemases and mcr-positive isolates might have emerged and distributed in fresh vegetables around the world, especially in China. However, isolates co-harboring mcr and carbapenemases-encoding genes have not been isolated in those previous studies. Considering the widespread of mcr and carbapenemases-encoding genes in China, it is crucial to identify mcr-positive CRE in fresh vegetables. In this study, we identified two mcr-positive E. coli strains producing NDM-5/9, recovered from fresh vegetables in China and the characteristics of resistance plasmids were also analyzed.
Materials and methods
Samples and identification of Enterobacteriaceae co-harboring mcr and carbapenemases genes
Seventeen different types of fresh vegetables from 72 supermarkets and farmer’s markets in 29 cities or districts of 10 provinces (Shandong, Shanghai, Beijing, Hubei, Henan, Heilongjiang, Yunan, Tianjin, Shanxi and Jiangsu) in China were purchased between May 2017 and December 2018. In total, 712 fresh vegetable samples were collected, including cucumber (n=125), tomato (n=114), romaine lettuce (n=76), curly endive (n=53), green pepper (n=50), coriander (n=50), leaf rape (n=49), spinach (n=49), mungbean sprouts (n=35), chili pepper (n=28), leaf lettuce (n=20), soybean sprouts (n=18), pakchoi (n=17), garland chrysanthemum (n=10), carrot (n=9), green shallots (n=5) and eggplant (n=4). These samples were processed with Mueller-Hinton (MH) broth co-harboring vancomycin (8 mg/L), colistin (2 mg/L) and meropenem (1.0 mg/L) to select Enterobacteriaceae carrying both carbapenemase and mcr genes using the similar protocol as previously reported.20 The MH broth with survived bacteria was diluted in series of 1:10 and 100 μL appropriate dilution was spread onto MH agar plates supplemented with both colistin (2 mg/L) and meropenem (1.0 mg/L). Presumptive Enterobacteriaceae colonies on the MH plate were selected for screening the carbapenemase-encoding genes (blaNDM, blaKPC, blaIMP, blaVIM, blaSPM, blaAIM, blaDIM, blaGIM, blaSIM, blaBIC and blaOXA-48) using primers previously described,25 and the presence of mcr (mcr-1, mcr-2, mcr-3, mcr-4, mcr-5, mcr-6, mcr-7 and mcr-8) was also determined.1 mcr-positive isolates carrying carbapenemase-encoding genes were identified by rpoB sequence analysis.26
Antimicrobial susceptibility testing
Susceptibility testing to 17 antimicrobial agents was determined by the agar dilution method according to the Clinical and Laboratory Standards Institute (CLSI) criteria.27 The 17 antimicrobial agents were the following ones: cefotaxime, ceftiofur, meropenem, ampicillin, enrofloxacin, ciprofloxacin, levofloxacin, nalidixic acid, streptomycin, amikacin, gentamicin, kanamycin, doxycycline, tetracycline, tigecycline, florfenicol and fosfomycin. The results except tigecycline were interpreted according to the CLSI breakpoints.27 The MIC method for colistin and breakpoints for colistin and tigecycline were recommended by the 2017 EUCAST (http://www.eucast.org/clinical_breakpoints/).
MLST and PFGE typing
MLST28 of the E. coli isolates carrying both carbapenemase and mcr genes in this study was performed to compare with such isolates of other origins. Clonal relationships of isolates in this study and those with the same MLST types from other sources were also investigated by PFGE using XbaI enzyme as previously described.29 The XbaI-digested DNA of Salmonella Braenderup strain H9812 was used as a reference.
Plasmid conjugation and incompatibility typing
Plasmid conjugation experiment was performed between the mcr-positive isolates carrying carbapenemase and streptomycin-resistant recipient E. coli C600 using the broth-mating method.30 mcr-positive transconjugants were selected on eosin methylene blue agar plates containing both streptomycin (2000 mg/L) and colistin (2 mg/L), while MacConkey agar plates supplemented with both streptomycin (2000 mg/L) and meropenem (0.8 mg/L) were used to select transconjugants with carbapenemase genes. Antimicrobial susceptibility testing and PCRs mentioned above were subsequently performed to confirm the transconjugants, followed by Enterobacterial repetitive intergenic consensus PCR as previously described.31 Incompatibility (Inc) groups of plasmids within the transconjugants were assigned by the PCR-based replicon typing method,32 and the IncX and IncI2 replicons were also detected as previously described.33,34
Plasmid analysis
To analyze the location of mcr and carbapenemase genes, S1 nuclease-PFGE and Southern Hybridization blot were performed twice on the transconjugants and their donors. E. coli C600 lacking plasmid was used to confirm the specific for mcr or carbapenemase gene probe. To determine whether the blaNDM/mcr-1-bearing plasmids in this study were similar to the reported plasmids of other origins, total genomic DNA (including the chromosome and corresponding plasmid) from the two mcr-1-positive transconjugants and two blaNDM-positive transconjugants were extracted and sequenced using Illumina HiSeq PE150, respectively. After assembling the sequence reads and cleaning out E. coli C600 chromosomal DNA sequences, plasmid contigs were obtained. All plasmids in this study were then subjected to PlasmidFinder 2.0 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) and ResFinder 3.1 (https://cge.cbs.dtu.dk/services/ResFinder/) to analyze the plasmid replicons and antimicrobial resistance genes, respectively. Functional annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline server, and the BLASTn implemented in software BRIG was used for sequence comparison.35
Results and discussion
Among the 712 fresh vegetable samples collected in 10 provinces from China in this study, two isolates from leaf rape and spinach in two supermarkets of Shandong province, respectively, carried both blaNDM-5/9 and mcr-1. To our knowledge, this is the first report of isolates co-carrying blaNDM-5/9 and mcr-1 in fresh vegetables.
The two isolates were designated 690 and 701 here. rpoB sequence analysis showed that both isolates were E. coli. Isolate 690 concomitantly harbored blaNDM-5 and mcr-1, while 701 carried both blaNDM-9 and mcr-1 (Table 1). Both isolates showed resistance to all beta-lactams, tetracyclines, fluoroquinolones, fosfomycin and colistin tested, which were therapeutic agents in clinics in many countries.36 Notably, both isolates remained susceptible to amikacin and tigecycline, similar to the E. coli isolates producing both NDM and MCR-1 from humans in China.9
 |
Table 1 Characteristics of the two E. coli strains 690 and 701 from fresh vegetables and their transconjugants with plasmids either harboring mcr-1 or blaNDM-5/-9 |
MLST analysis showed that isolates 690 and 701 belonged to ST156 and ST2847, respectively. ST156 and ST2847 were also found in E. coli isolates co-carrying mcr-1 and blaNDM from avian in Shandong of China in 2015, in our previous study,12 suggesting that the MCR-1-NDM producers in vegetables might have originated from animals because vegetables might be fertilized with manure and wastewater from livestock. In fact, the final effluent applied to farmland in the piggery wastewater treatment system in China has been proved to contain considerable amounts of blaNDM and mcr-1,37 which will further support our hypothesis. We then performed PFGE using XbaI enzyme to compare the clonal relationships of isolates in this study and isolates of ST156 and ST2847 types from avian we previously reported.12 The results showed that the ST156 isolate 690 was different from the five ST156 isolates from avian, and isolate 701 of ST2847 was also different from the two ST2847 isolates of avian origin (Figure 1), suggesting that such isolates in vegetables in this study were not directly derived from the avian feces we studied previously and more E. coli isolates co-carrying mcr-1 and blaNDM from other animal farms should be investigated in the future. Notably, ST156 type of E. coli isolates producing NDM-5 and isolate carrying mcr-1 were also found in human from China in 20169 and human from Brazil in 2016,38 respectively, while ST2847 E. coli isolated from a patient in Hong Kong in 2004 was found to carry both blaCTX-M-65 and fosA3.39 Thus, the presence of ST156 and ST2847 E. coli isolates producing both NDM-5/9 and MCR-1 in vegetables still represents a threat to human health.
 |
Figure 1 XbaI-PFGE patterns of isolates in this study and isolates of ST156 and ST2847 types co-carrying mcr-1 and blaNDM from avian. |
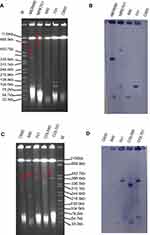
For both isolates, we obtained two different transconjugants harboring mcr-1 or blaNDM-5/9, respectively (Table 1). fosA3, conferring resistance to fosfomycin, was found in blaNDM-9-positive transconjugants MER701 using primers described previously,40 while no other resistances were co-transferred with colistin/meropenem resistance in the other 3 transconjugants. Interestingly, there were additional bigger bands marked with arrows in all 4 transconjugants and some donors in the S1-PFGE (Figure 2A and C) and these bands could be also hybridized with the corresponding mcr-1/blaNDM probe (Figure 2B and D), although these experiments were performed several times. These bigger bands were the portion of the mcr-1/blaNDM-5/9-carrying plasmids not exposed to S1 nuclease in the S1-PFGE experiment, according to the findings of the previous study in which the S1-PFGE method was established.41 Thus, all the 4 transconjugants carried only one plasmid and mcr-1 was located on IncX4 type plasmid of ~33 kb in COL690, while COL701 carried IncI2 type ~60 kb plasmid harboring mcr-1 (Figure 2C and D). blaNDM-5 was on IncX3 plasmid of ~40 kb in transconjugant MER690, however, blaNDM-9 and fosA3 were on the same ~110 kb plasmid which was untypable in MER701 (Figure 2A and B and Table 1).
From the results of plasmid sequences, both mcr-1-bearing plasmids did not carry any other antibiotic resistance gene besides mcr-1 and this could account for the resistance phenotypes of transconjugants COL690 and COL701 (Table 1). Comparison of the pCOL690T (accession no. VMKQ00000000) to several previously reported ~33-kb IncX4 plasmids showed that it aligned very well to pCSZ4 (KX711706) (100% in coverage and 99% in identity) from E. coli of pork origin in China (Figure 3A). Notably, pCOL690T (VMKQ00000000) from leaf rape in this study was also highly similar to plasmids pKP15450-MCR-1 (MH715959) from clinical Klebsiella pneumoniae in Taiwan, pmcr1_IncX4 (KU761327) from clinical K. pneumoniae in China and pNG14043 (KY120364) from clinical Salmonella Typhimurium in Taiwan (Figure 3A). Furthermore, the pCOL690T in the mcr-1-positive NDM-5-producing E. coli in this study also showed high similarity to plasmid pMCR-1-IHIT35346 (KX894453) from E. coli co-producing OXA-181-carbapenemase and mcr-1 of pig origin in Germany. Plasmid pCOL701T (VMKR00000000) from E. coli of spinach in this study aligned well to IncI2 plasmid pP111 (KY120365) (99% in coverage and 100% in identity) from S. Typhimurium of pig in Taiwan, pHNGDF93 (MF978388) from fish E. coli and p1106-IncI2 (MG825374) from E. coli of chicken in China (Figure 3B). Notably, the pCOL701T (VMKR00000000) in NDM-9-producing E. coli in this study was also highly similar to p5CRE51-MCR-1 (CP021176) from clinical E. coli co-producing NDM-9 and MCR-1 in Taiwan. All these findings suggested that highly similar IncX4 or IncI2 plasmids have disseminated mcr-1 among different Enterobacteriaceae species in food and animals around the world, and these plasmids can spread to carbapenemase-producing clinical isolates to threaten human health.
The pMER690T plasmid (VMKS00000000) did not carry any other resistance gene besides blaNDM-5 (Figure 3C), and it aligned very well to IncX3 plasmid p977-NDM (MG825382) (100% in coverage and 99% in identity) from E. coli of pork origin in China (Figure 3C). Notably, the pMER690T from leaf rape in this study was also highly similar to plasmids pCREC-591_4 (CP024825) from clinical E. coli in South Korea, and pJEG027 (KM400601) from clinical K. pneumoniae in Australia. Interestingly, the pMER690T (VMKS00000000) in the mcr-1-positive E. coli producing NDM-5 in this study also showed high similarity to plasmid pVH1 (CP028705) from E. coli of cucumber in China in our previous report,24 in which only CRE isolates were isolated. Replicon untypable plasmid pMER701T (VMKT00000000) carried resistance genes fosA3, dfrA12, aadA2, sul1, bleMBL, blaNDM-9, and mph(A), which were all centralized in the multidrug resistance region (Figure 3D). pMER701T (VMKT00000000) in this study aligned well to plasmids pHNTH02-1 (MG196294) (100% in coverage and 99% in identity), p1106-NDM (MG825375) and p92944-NDM (MG838206), and these three plasmids were from E. coli of retail meat, chicken and human, respectively, in China (Figure 3D). These findings suggested that pMER690T- and pMER701T-like plasmids have been disseminated among different Enterobacteriaceae species of various origins around the world, especially China.
Conclusion
In summary, we reported for the first time two clonally unrelated E. coli harboring both blaNDM-5/9 and mcr-1 in fresh vegetables in China. The dissemination of mcr-1 was mediated by IncX4 or IncI2 plasmid, while blaNDM-5/9 was on IncX3 or untypable plasmid. All the plasmids in this study were highly similar to the plasmids from animals and clinical isolates in various countries. The emergence of mcr-1-positive bacteria producing NDM in fresh vegetables is alarming and constitutes a food safety issue. Further investigations are required for monitoring such organisms in fresh vegetables to ensure food safety in China and other countries.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (number 31502122), the Scientific and Technological Projects of Qingdao (19-6-1-94-nsh) and the Advanced Talents Foundation of Qingdao Agricultural University (number 663/1115014).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi:10.1038/s41426-018-0124-z
2. Roschanski N, Fischer J, Falgenhauer L, et al. Retrospective analysis of bacterial cultures sampled in german chicken-fattening farms during the years 2011-2012 revealed additional VIM-1 carbapenemase-producing Escherichia coli and a serologically rough Salmonella enterica serovar infantis. Front Microbiol. 2018;9:538. doi:10.3389/fmicb.2018.00538
3. Li Y, Sun QL, Shen Y, et al. Rapid increase in prevalence of carbapenem-resistant Enterobacteriaceae (CRE) and emergence of colistin resistance gene mcr-1 in CRE in a hospital in Henan, China. J Clin Microbiol. 2018;56(4). doi:10.1128/JCM.01932-17
4. Huang TD, Bogaerts P, Berhin C, et al. Increasing proportion of carbapenemase-producing Enterobacteriaceae and emergence of a MCR-1 producer through a multicentric study among hospital-based and private laboratories in Belgium from September to November 2015. Euro Surveill. 2017;22(19):30530. doi:10.2807/1560-7917.ES.2017.22.19.30530
5. Mediavilla JR, Patrawalla A, Chen L, et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio. 2016;7(4). doi:10.1128/mBio.01191-16
6. Delgado-Blas JF, Ovejero CM, Abadia-Patino L, Gonzalez-Zorn B. Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob Agents Chemother. 2016;60(10):6356–6358. doi:10.1128/AAC.01319-16
7. Feng S, Shen C, Chen H, et al. Co-production of MCR-1 and NDM-5 in Escherichia coli isolated from a colonization case of inpatient. Infect Drug Resist. 2018;11:1157–1161. doi:10.2147/IDR.S171164
8. Quan J, Li X, Chen Y, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. doi:10.1016/S1473-3099(16)30528-X
9. Shen Z, Hu Y, Sun Q, et al. Emerging carriage of NDM-5 and MCR-1 in Escherichia coli from healthy people in multiple regions in china: a cross sectional observational study. EClinicalMedicine. 2018;6:11–20. doi:10.1016/j.eclinm.2018.11.003
10. Yao X, Doi Y, Zeng L, Lv L, Liu JH. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 2016;16:288–289. doi:10.1016/S1473-3099(16)00057-8
11. Wang Y, Zhang R, Li J, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2017;2:16260. doi:10.1038/nmicrobiol.2016.251
12. Liu BT, Song FJ, Zou M, Zhang QD, Shan H. High incidence of Escherichia coli strains coharboring mcr-1 and blaNDM from chickens. Antimicrob Agents Chemother. 2017;61(3). doi:10.1128/AAC.02347-16
13. Callejon RM, Rodriguez-Naranjo MI, Ubeda C, Hornedo-Ortega R, Garcia-Parrilla MC, Troncoso AM. Reported foodborne outbreaks due to fresh produce in the United States and European Union: trends and causes. Foodborne Pathog Dis. 2015;12(1):32–38. doi:10.1089/fpd.2014.1821
14. Jung Y, Jang H, Matthews KR. Effect of the food production chain from farm practices to vegetable processing on outbreak incidence. Microb Biotechnol. 2014;7(6):517–527. doi:10.1111/1751-7915.12178
15. van Hoek AH, Veenman C, van Overbeek WM, Lynch G, de Roda Husman AM, Blaak H. Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int J Food Microbiol. 2015;204:1–8. doi:10.1016/j.ijfoodmicro.2015.03.014
16. Zurfluh K, Nuesch-Inderbinen M, Morach M, Zihler Berner A, Hachler H, Stephan R. Extended-spectrum-beta-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl Environ Microbiol. 2015;81(9):3115–3120. doi:10.1128/AEM.00258-15
17. Jones-Dias D, Manageiro V, Ferreira E, et al. Architecture of class 1, 2, and 3 integrons from gram negative bacteria recovered among fruits and vegetables. Front Microbiol. 2016;7:1400. doi:10.3389/fmicb.2016.01400
18. Zurfuh K, Poirel L, Nordmann P, Nuesch-Inderbinen M, Hachler H, Stephan R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-beta-lactamase-producing enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother. 2016;60(4):2594–2595. doi:10.1128/AAC.00066-16
19. Luo J, Yao X, Lv L, et al. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegeTAbles in China. Antimicrob Agents Chemother. 2017;61(10). doi:10.1128/AAC.01139-17
20. Liu BT, Li X, Zhang Q, Shan H, Zou M, Song FJ. Colistin-resistant mcr-positive enterobacteriaceae in fresh vegetables, an increasing infectious threat in China. Int J Antimicrob Agents. 2019;54(1):89–94. doi:10.1016/j.ijantimicag.2019.04.013
21. Zurfluh K, Poirel L, Nordmann P, Klumpp J, Stephan R. First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob Resist Infect Control. 2015;4:38. doi:10.1186/s13756-015-0080-5
22. Touati A, Mairi A, Baloul Y, et al. First detection of Klebsiella pneumoniae producing OXA-48 in fresh vegetables from Bejaia city, Algeria. J Glob Antimicrob Resist. 2017;9:17–18. doi:10.1016/j.jgar.2017.02.006
23. Wang J, Yao X, Luo J, Lv L, Zeng Z, Liu JH. Emergence of Escherichia coli co-producing NDM-1 and KPC-2 carbapenemases from a retail vegetable, China. J Antimicrob Chemother. 2018;73(1):252–254. doi:10.1093/jac/dkx335
24. Liu BT, Zhang XY, Wan SW, Hao JJ, Jiang RD, Song FJ. Characteristics of carbapenem-resistant enterobacteriaceae in ready-to-eat vegetables in China. Front Microbiol. 2018;9:1147. doi:10.3389/fmicb.2018.01147
25. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
26. Mollet C, Drancourt M, Raoult D. rpoB sequence analysis as a novel basis for bacterial identification. Mol Microbiol. 1997;26(5):1005–1011. doi:10.1046/j.1365-2958.1997.6382009.x
27. CLSI. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. CLSI document M100-S25. Wayne (PA): Clinical and Laboratory Standards Institute; 2015.
28. Wirth T, Falush D, Lan R, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136–1151. doi:10.1111/j.1365-2958.2006.05172.x
29. Saenz Y, Brinas L, Dominguez E, et al. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother. 2004;48(10):3996–4001. doi:10.1128/AAC.48.10.3996-4001.2004
30. Chen L, Chen ZL, Liu JH, Zeng ZL, Ma JY, Jiang HX. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J Antimicrob Chemother. 2007;59(5):880–885. doi:10.1093/jac/dkm065
31. Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA-sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19(24):6823–6831. doi:10.1093/nar/19.24.6823
32. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi:10.1016/j.mimet.2005.03.018
33. Johnson TJ, Bielak EM, Fortini D, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68(1):43–50. doi:10.1016/j.plasmid.2012.03.001
34. Chen L, Chavda KD, Al Laham N, et al. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother. 2013;57(10):5019–5025. doi:10.1128/AAC.01397-13
35. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi:10.1186/1471-2164-12-402
36. Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum beta-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect Dis. 2010;10(1):43–50. doi:10.1016/S1473-3099(09)70325-1
37. Yang F, Gu Y, Zhou J, Zhang K. Swine waste: a reservoir of high-risk blaNDM and mcr-1. Sci Total Environ. 2019;683:308–316. doi:10.1016/j.scitotenv.2019.05.251
38. Rossi F, Girardello R, Morais C, et al. Plasmid-mediated mcr-1 in carbapenem-susceptible Escherichia coli ST156 causing a blood infection: an unnoticeable spread of colistin resistance in Brazil? Clinics (Sao Paulo). 2017;72(10):642–644. doi:10.6061/clinics/2017(10)09
39. Ho PL, Chan J, Lo WU, et al. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. J Med Microbiol. 2013;62(Pt 11):1707–1713. doi:10.1099/jmm.0.062653-0
40. Hou J, Huang X, Deng Y, et al. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M beta-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob Agents Chemother. 2012;56(4):2135–2138. doi:10.1128/AAC.05104-11
41. Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226(2):235–240. doi:10.1006/abio.1995.1220
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.