Back to Journals » Infection and Drug Resistance » Volume 13
Emergence of NDM-5-Producing Carbapenem-Resistant Klebsiella pneumoniae and SIM-Producing Hypervirulent Klebsiella pneumoniae Isolated from Aseptic Body Fluid in a Large Tertiary Hospital, 2017–2018: Genetic Traits of blaNDM-Like and blaSIM-Like Genes as Determined by NGS
Authors Li Q, Zhu J, Kang J, Song Y, Yin D, Guo Q, Song J, Zhang Y, Wang S , Duan J
Received 11 May 2020
Accepted for publication 28 July 2020
Published 4 September 2020 Volume 2020:13 Pages 3075—3089
DOI https://doi.org/10.2147/IDR.S261117
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Qi Li,1,* Jiaying Zhu,1,* Jianbang Kang,2 Yan Song,2 Donghong Yin,2 Qian Guo,2 Junli Song,2 Yan Zhang,3 Shuyun Wang,2 Jinju Duan2
1Department of Pharmacy, School of Pharmacy, Shanxi Medical University, Taiyuan, Shanxi, People’s Republic of China; 2Department of Pharmacy, Second Hospital of Shanxi Medical University, Taiyuan, Shanxi, People’s Republic of China; 3Department of Chief Executive, Willingmed Technology (Beijing) Co., Ltd, Beijing, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shuyun Wang; Jinju Duan
Department of Pharmacy, Second Hospital of Shanxi Medical University, No. 382, Wuyi Road, Xinghualing District, Taiyuan, Shanxi, People’s Republic of China
Tel +86 351 3365713
Email [email protected]; [email protected]
Purpose: To characterize the clinical, resistance, and virulence features of carbapenem-resistant Klebsiella pneumonaie (CRKP) and hypervirulent Klebsiella pneumoniae (hvKP) and also provide an effective selection of drug in CRKP and hvKP treatment.
Materials and Methods: Twelve strains were collected and investigated these isolates for their antimicrobial susceptibility and molecular features. Resistance mechanisms, virulence-associated genes, multilocus sequence typing (MLST), and serotypes were detected by PCR and sequencing. Next general sequencing (NGS) was carried out to determine the features of carbapenem resistance and virulence. The synergistic activity of tigecycline–imipenem (TGC+IPM), tigecycline–meropenem (TGC+MEM), and tigecycline–aztreonam (TGC+ATM) combinations were performed by microdilution checkerboard method.
Results: Eleven CRKP and one hvKP strains were collected. All strains showed highly sensitive rates to tigecycline (TGC) and amikacin (AMK). NDM (33.3%, 4/12) was the main resistance mechanism and MLST assigned 3 of them to ST11. CTX-M-producing (n = 1) and KPC-2-producing (n = 1) isolates belonged to ST147 and ST11, respectively. The MICs of ATM and quinolones in NDM-1 CRKP and NDM-5 CRKP strains were different. The serotype of the majority strains was KL22KL137 (58.3%, 7/12), hvKP stain belonged to K64. CRKP strains harbored plasmid-mediated quinolone resistance genes (oqxA, oqxB, qnrS, qnrB), β-lactams (blaCTX-M-3), aminoglycosides, type I and type III fimbriae genes, siderophore genes, and transporter and pumps. SIM-producing ST1764 K64 showed typical features of hvKP, showing hypermucoviscosity phenotype. The virulence genes, including rmpA2, alls and aerobactin genes, linked to hvKP, were found in ST1764 hvKP. hvKP was sensitive to quinolone; also, oqxA gene was detected. All TGC combinations showed highly synergistic effects and TGC+IPM was more effective treatment.
Conclusion: We first identified the NDM-5-producing ST690 CRKP and SIM-producing ST1764 hvKP strains in Shanxi province. Tigecycline-carbapenem combinations were available treatments for CRKP.
Keywords: Klebsiella pneumoniae, hypermucoviscous, blaNDM-5, ST1764, tigecycline, synergistic effect
Introduction
Klebsiella pneumoniae (KP) is a pathogenic bacterium, which causes infections in a variety of sites including the urinary tract, lungs, liver and bloodstream.1,2 KP has earned notoriety as resistance to “last resort” antimicrobials: carbapenems.3,4 From the China Network Antibacterial Surveillance Center, the carbapenem antibiotics usage density (AUD, defined as DDDs/per 100 patient-days) increased from 2.04 in 2013 to 3.38 in 2017. According to CHINET, the rates of KP resistance to imipenem and meropenem increased from 10.3% and 14.1% in 2013 to 25% and 26.3% in 2018, respectively. Carbapenem-resistant Klebsiella pneumoniae (CRKP) has become a crucial threat to public health.2,5
The expression of carbapenemases is one of the main carbapenem-resistance mechanisms in KP.6 Carbapenemases include three types: class A (blaKPC, blaGES and blaIMI), class B (blaNDM, blaIMP, blaVIM, and blaSIM) and class D (blaOXA-48 like).7 In China, carbapenem resistance has mainly resulted from the dissemination of KPCs, especially blaKPC-2.8 A study identified and reported the first KPC-positive KP strain in 2004 from Zhejiang province.9 Among class B, New Delhi metallo-β-lactamase (NDM) is the main resistance gene that can hydrolyze all β-lactams antibiotics except monobactams.10 Hitherto, more than 20 different KPC variants and 21 different NDM variants have been observed worldwide.10–12 NDM-1 enzyme hydrolyzes carbapenems and is not targeted by beta-lactamase inhibitors.5 In 2011, NDM-5 was first identified in Escherichia coli isolates in the UK.13 This enzyme has a higher resistance to carbapenems and extended-spectrum cephalosporins in KP.10 Recent researches have reported the outbreaks of NDM-5-producing CRKP in China.10,13 Thus, NDM-producing CRKP poses a great challenge for clinical treatment.10 From the standpoint of geographical distribution, KPC-2-producing KP has been reported in certain areas in China,14,15 while another study showed that the clinically isolated CRKP strains in Northwestern China produced a high level of NDM enzymes.16 Therefore, analyzing the local data can aid in optimizing the future strategies of antimicrobial usage and providing meaningful guidance to the clinical practice.17
The virulence factors of KP lead to high mortality and enhance the difficulty of treatment. Most of the studies classified the virulence genes as four major classes, including type I and type III fimbriae, lipopolysaccharide, siderophore iron uptake systems, and a polysaccharide capsule.18 Hypermucoviscous (HM) is a feature of Hypervirulent Klebsiella pneumoniae (hvKP) strains. Furthermore, hvKP isolates secrete more siderophores, such as aerobactin.19 A study showed that only about 6% of KP strains expressed aerobactin, yet aerobactin present in 93% to 100% of hvKP isolates.6
The present study aimed to conduct an in-depth molecular characterization of CRKP and hvKP isolated from aseptic body fluid and provide a favorable basis for the combined treatment of CRKP infection.
Materials and Methods
Bacterial Collection
From September 28, 2017, to December 14, 2018, 12 non-duplicated clinical CRKP isolates originated from different aseptic body fluid: blood (n = 7, 63.6%, 7/11), cerebrospinal fluid (n = 2, 18.2%, 2/11), pus (n = 1, 9.1%, 1/11), and puncture fluid (n = 1, 9.1%, 1/11) were collected from the Second Hospital of Shanxi medical university. During the study, CRKP isolate was defined as a clinical isolate with non-susceptibility to carbapenem (imipenem, meropenem, or ertapenem), in accordance with the breakpoints of Clinical and Laboratory Standards Institute (CLSI-2019) guidelines.20 All isolates were stored at −80°C for antimicrobial susceptibility testing and investigation of resistance mechanisms and virulence genes.
Clinical Data Collection
The clinical information was collected from the medical records of each patient, including patient demographics, underlying medical conditions, invasive operation during hospitalization, antimicrobial therapy, and outcomes.
This study was reviewed and approved by the research ethics committee of the Second Hospital of Shanxi Medical University (2019 YX-181). We had hidden the patient’s information. The patients’ written informed consent was exempt. This study was also in line with the guidelines outlined in the Declaration of Helsinki. Our data were expressed as means ± standard deviation of the mean (SD).
Antimicrobial Susceptibility Testing
All isolates were reidentified by an automated VITEK-2 compact system (BioMerieux Italia S.p.A) and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDITOF-MS) (Bruker Daltonik, Bremen, Germany). Antimicrobial susceptibility was evaluated by the agar dilution and broth microdilution methods according to the CLSI-2019 guidelines. The minimum inhibitory concentration (MIC) breakpoints were interpreted according to CLSI-2019.20 The breakpoints proposed by the Food and Drug Administration (FDA) standard (FDA-2016) guidelines were used for tigecycline (TGC).21 The following antibiotics were tested: meropenem (MEM), imipenem (IPM), ertapenem (ETP), ceftriaxone (CRO), ceftazidime (CAZ), cefotaxime (CTX), cefepime (FEP), aztreonam (ATM), cefoperazone/sulbactam (CSL), piperacillin/tazobactam (TZP), ciprofloxacin (CIP), levofloxacin (LEV), fosfomycin (FOS), chloramphenicol (CHL), amikacin (AMK), and tigecycline (TGC).
Detection of Carbapenemase Phenotypes and Resistance Mechanism
According to the CLSI-2016 and CLSI-2019 recommendations,20,22 the Modified Carbapenem Inactivation Method (mCIM) test, ethylenediaminetetraacetic acid-Modified Carbapenem Inactivation Method (eCIM), and Modified Hodge Test (MHT) were used for phenotypic detection of carbapenemase. The polymerase chain reaction (PCR) was used to detect the genes encoding carbapenemases (blaKPC, blaGES, blaIMI, blaNDM, blaIMP, blaVIM, blaSIM and blaOXA-48), ESBLs, AmpC β-lactamases (blaCTX-M-2,4–7, blaCTX-M-9,13–14,16–19, blaCTX-M-1,3, 10–12,15, DHA, ACT and CMY), and the colistin resistance gene mcr-1, as previously described.16
Multilocus Sequence Typing (MLST)
We performed MLST as described on the Pasteur Institute MLST website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). The sequences of seven housekeeping genes were compared with those in the MLST databases (Institute Pasteur MLST Database; https://bigsdb.pasteur.fr/Klebsiella).
Phenotypical Identification and Definition of hvKP
We used the string test to detect hypermucoviscosity phenotype as described previously.23 The positive for the string test was defined as the formation of a viscous string exceeding 5 mm in length, as previously described.24 Multiplex PCR for virulence genes including iucA, iutA, rmpA, rmpA2 and iroN was performed after phenotypic characterization as previously described.25 The KP isolates with positive result were defined as HMKP. The HMKP isolates with positive results of rmpA, rmpA2 and iroN genes were defined as hvKP in this study.
Identification of Serotypes
The serotypes were detected by wzi gene.26 The sequences of wzi gene were compared with the wzi databases (https://bigsdb.pasteur.fr/Klebsiella). Positive PCR products were visualized by agarose gel electrophoresis, purified with a QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA) and sequenced by Sanger sequencing on an ABI PRISM 3730XL system (Applied Biosystems, Foster City, CA, USA).
Next Generation Sequencing (NGS)
Three representative isolates were screened out. The genomes of three isolates were fully sequenced by Ion S5 plus (Thermo fisher) with 420 flows, which obtained read lengths ranging from 100bp to 350bp after read filtering. De novo assembly was performed using SPAdes genome assembler. Antimicrobial resistance and virulence genes were screened using CARD and VFDB tools with an identity threshold of 97%.27
Tigecycline Combinations Checkboard Assay
Synergistic activity of tigecycline–imipenem (TGC+IPM), tigecycline–meropenem (TGC+MEM), and tigecycline–aztreonam (TGC+ATM) combinations were performed by microdilution checkerboard method using 96-well microtiter plates (Corning Corporation, America). The concentration ranges were based on the MICs determined above. Concentrations of each antimicrobial tested in combination range from 1/32× MIC to 4× MIC. The volume of mixture antibiotics in each well was 100 μL. Then, each well was inoculated with 100 μL of a 5 ×105 CFU/mL suspension of the test CRKP isolates in a final volume of 200 μL.
The effects of antibiotics in combination were quantified by the fractional inhibitory concentration (FIC) index. The FIC index ≤0.5 was interpreted as synergy, >0.5 to ≤4.0 as additive, and >4.0 as antagonism.28 The cation-adjusted Mueller-Hinton broth (CAMHB) served as negative control (Hepebio, Qingdao Hope Biotechnology, Qingdao, Shandong, China) and bacterial suspension served as positive control.
Results
Clinic and Demographic Characteristics of Patients Infected with CRKP and hvKP
In the present study, a total of 12 KP isolates were collected from aseptic body fluid from September 2017 to December 2018. Eleven of them were resistant to carbapenems and were termed as CRKP with a carriage rate of 91.67% (11/12). The remaining one isolate was sensitive to carbapenem and was further defined as hvKP.
The hvKP strain originated from the hydropericardium of a 48-year-old male in February 2018. This patient had pyogenic liver abscesses (PLA) disease and developed septic shock. Although the patient received antibiotics, the specific medication information was unclear. Eventually, the patient was deceased (Table 1).
 |
Table 1 Clinical Characteristics of Patients Infected with CRKP and hvKP |
The median age of CRKP patients was 60 years (range 44–85 years). The majority of CRKP isolates were isolated from the intensive care unit (ICU) (n = 5, 45.5%, 5/11). Seven (63.6%, 7/11) patients had history of ICU (Table 1). The most common sites of infection were pneumoniae infection (n = 6, 54.5%, 6/11), followed by bloodstream infection (n = 4, 33.3%, 4/11). (Table 2) 83.3% (5/6) of pneumoniae patients had other co-infection, such as bloodstream infection (n =2, 33.3%, 2/6), catheter related bloodstream infection (n = 1, 16.7%, 1/6), intracranial infection (n = 1, 16.7%, 1/6), and pus infection (n = 1, 16.7%, 1/6). The proportion of immune deficiency was 81.8% (n = 9, 9/11). (Table 2)
 |
Table 2 Demographics of Patients Infected with CRKP and hvKP |
All patients underwent invasive procedures during the hospitalization. The mortality of this study was 58.3% (7/12).
Therapeutic Regimens and Prognostic of Patients with CRKP Infection
Carbapenems were administered as monotherapy for 6 patients (54.5%, 6/11) and 5 died. Tigecycline was given as monotherapy for 4 patients (36.4%, 4/11) and three died. One patient received the combination of TGC with carbapenem. And this patient was discharged. All the patients had received empiric therapies prior to the first positive culture. 81.8% (n = 9, 9/11) of patients had received combination therapy during hospitalization. 66.7% of patients (n = 6, 6/9) received the combination of TGC with non-carbapenems therapies and four patients died. 22.2% (n = 2, 2/9) of the CRKP patients were treated with carbapenems combined with other antibiotics (except TGC) therapies and one patient died. The detailed information is listed in Figure 1.
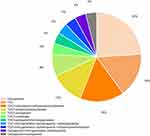 |
Figure 1 Distribution of therapeutic regimens of patients with CRKP infection. |
Antimicrobial Susceptibility
The results of the susceptibility testing revealed that the most highly resistant rates of 11 CRKP isolates were third- or fourth-generation cephalosporins (100%) and CSL (100%, each), followed by IPM (90.9%, 10/11), MEM (90.9%, 10/11), ATM (90.9%, 10/11), ETP (81.8%, 9/11), TZP (81.8%, 9/11) and quinolones (81.8%, 9/11) (Table 3). More than 90.9% of the CRKP isolates had high MICs (≥32 mg/L) for β-lactam combination agents (ATM, CSL and TZP). All CRKP isolates were susceptible to AMK (100%), followed by TGC (63.6%, 7/11), and FOS (54.5%, 6/11). The hvKP isolate showed lower resistance to antibiotics, especially carbapenem (100%). It was resistant to TGC (MIC = 8 mg/L) and AMK (MIC = 1 mg/L) (Table 3).
 |
Table 3 Antimicrobial Susceptibility Testing Results of 12 K. pneumoniae Isolates (Mg/L) |
Carbapenemase Phenotypes and Resistance Mechanism
66.7% (8/12) of the KP isolates were positive for the mCIM and eCIM tests. Of the 11 CRKP isolates, the most prevalent resistance mechanism was NDM type (n = 4, 36.4%, 4/11), followed by ESBLs type (CTX-M-3 and CTX-M-14) (n =2, 18.2%, 2/11), and KPC type (n = 1, 9.1%, 1/11). Two CRKP isolates were found to express two types of resistance genes (NDM-5/SIM and CTX-M-14/KPC-2). The resistance mechanism of hvKP was SIM type, but not in accordance with the result of carbapenemase phenotypes assay, whereas 41.7% (5/12) of isolates were not found any resistance genes. The resistance genes OXA-48, AmpC and colistin resistance genes mcr-1 were not detected in all isolates (Table 4).
 |
Table 4 Distribution of Resistance Mechanism and Virulence Genes Harbored by CRKP and hvKP Strains |
Distribution of MICs with Different Carbapenemases (NDM and KPC-2/CTX-M-14)
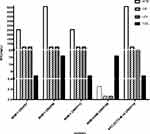
We identified 3 carbapenemases in 12 isolates. For 3 NDM-1-producing isolates, the MIC of ATM ranges from 256 to 512 mg/L. NDM-1-producing isolates were resistant to quinolones (MIC ≥ 64 mg/L). In contrast to NDM-1-producing isolates, the NDM-5-producing isolate had a MIC < 0.25 mg/L for ATM. And it was sensitive to quinolones (MIC = 0.064 mg/L). NDM-5-producing CRKP had lower MIC of IPM and higher MIC of ETP than NDM-1-producing CRKP strains. Furthermore, the MEM, IPM, and ETP MICs for KPC-2/CTX-M-14 co-harboring isolate were 64 mg/L, 32 mg/L, 128 mg/L, respectively. And it had high MICs for most test antibiotics (≥32 mg/L) (Figure 2).
MLST Analysis
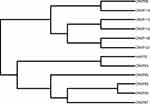
We have detected six sequence types (STs), ST11 (58.3%, 7/12) was the main type and detected in seven CRKP isolates. Other STs were also identified: ST147, ST690, ST7, ST524, and ST1764. They all accounted for 8.3% (1/12), respectively. Moreover, ST11 strains were detected from April 2018 to August 2018. hvKP isolate belonged to ST1764 (gapA_5, infB_3, mdh_1, pgi_1, phoE_9, rpoB_4, tonB_283), which is a three-locus variant of ST23 (gapA_2, infB_1, mdh_1, pgi_1, phoE_9, rpoB_4, tonB_12) (Table 4). From Figure 3, the ST11 (CRKP110) isolate had a higher affinity with ST11 (CRKP112) isolate, they were isolated from blood and cerebrospinal fluid, respectively. ST147 (CRKP58) isolate and ST11 (CRKP90) isolate shared the common ancestor.
Virulence-Associated Features and Serotypes Results
All isolates were performed the string test and only ST1764 was positive. The virulence genes, including iucA, rmpA, rmpA2, and iroN, were evaluated, and only CRKP95 ST11 and hvKP75 ST1764 were positive. We defined hvKP as hypermucoviscosity and PCR screening were positive. Intriguingly, seven of the isolates (58.3%, 7/12) belonged to serotype KL22KL37. 33.3% (4/12) of the isolates were typed as KL106, KL54, KL47, and KL64, while 8.3% (1/12) were not linked to any KL-type. Three different K-types were identified by wzi gene. 8.3% (n = 1, 1/12, each) of the isolates belonged to serotype K41, K47, and K64, respectively. We did not find any serotypes of nine isolates through wzi gene (75%, 9/12). (Table 4).
Distribution of Resistance Genes
The next-generation sequencing analysis was performed on three isolates (ST147 CTX-M-3-producing CRKP, ST11 NDM-1-producing CRKP, and ST1764 SIM-producing hvKP). In the two CRKP isolates, the presence of multiple genes encoding resistance to β-lactams (blaCTX-M-3), aminoglycosides [AAC (3)-IIb], quinolones (oqxAB, qnrS), macrolides [mphA, Mrx] were confirmed.
CTX-M-3-producing ST147 CRKP isolate carried the plasmid-mediated quinolone resistance qnrB gene and aadA6/aadA10 gene, which were resistant to aminoglycosides. Alarmingly, ST147 CRKP harboring genes conferred tigecycline resistance [tet (C)] and chloramphenicol resistance [floR]. NDM-1-producing ST11 CRKP isolate carried ESBLs gene (blaSHV-187). A total of seven different blaNDM genes were identified within the ST11 CRKP. We also found many resistance genes in hvKP isolate. SIM-producing ST1764 hvKP carried genes responsible for the quinolone, ESBLs, fosfomycin resistance genes (oqxA, blaSHV-71, FosA2). The efflux pumps genes (emrB, ramA) were observed in hvKP (Table 5).
 |
Table 5 The Distribution of Resistance Genes |
Furthermore, compared to the CARD, ST147 CRKP carried 16 blaCTX-M genes and 15 qnrB genes; ST11 CRKP carried 8 ade cluster genes, 16 blaCTX-M genes and 7 blaNDM genes. These findings were different with the CARD. Compared the resistance genes of ST1764 isolate with CARD, the presence of genes encoding resistance to β-lactams (blaSHV-71) and fosfomycin (FosA2) had a significant difference.
Distribution of Virulence Genes
Three isolates exhibited multiple virulence profiles, namely the type I fimbriae cluster (FimABCDEFH), the mannose-resistant Klebsiella-like (type III) fimbriae cluster (mrkABCDFHIJ), the siderophore genes (IroE, iroN, iutA, aerobactin encoding iucABCD, enterobactin genes), transporter and pumps (ABC transporter, RcsAB, RND efflux system, adeFGH efflux pump/transport autoinducer). The results of the virulence genes are listed in Table 6. ST1764 (hvKP) isolate had the highest number of virulence genes (n = 43), especially the adhesion gene (rmpA2), allantoin metabolism genes (alls) and iucA~D genes. Moreover, none of the ferric uptake system (kfuABC) genes were examined in these isolates, which has a strong association between the expression of this factor and hvKP strains.
 |
Table 6 The Distribution of Virulence Genes |
Compared with the VFDB, we had detected the UTP-glucose-1-phosphate uridylyltransferase subunit (GalF), fimbrial chaperone protein mrkB precursor, siderophore esterase IroE, phosphomannomutase, mannose-1-phosphate guanylyltransferase genes in our study. All isolates carried GalF, and the remaining genes account for 66.7%.
Synergistic Effects of Tigecycline with Other Antibiotics
Synergistic effects, additive effects and antagonism effects were detected in TGC combinations with the checkerboard test. The TGC+IPM, TGC+MEM, and TGC+ATM combinations decreased the MICs of TGC by 4~5-fold. The MICs of imipenem decreased by 4~5-fold. As for MEM, and ATM, except the MICs of one isolate decreased only 1-fold, the MICs of the remaining decreased by 4~5-fold and 5-fold, respectively. TGC+IPM, TGC+MEM, and TGC+ATM combinations were high synergistic against 100% (11/11), 90.1% (10/11), and 90% (9/10) of CRKP isolates, respectively. Moreover, antagonism was not observed in these isolates in either of the combinations. The results of the FICs are shown in Table 7.
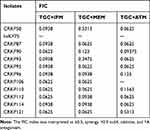 |
Table 7 The FICs of CRKP Isolates |
Discussion
Currently, CRKP strains are a major threat to public health worldwide.16 At the same time, the treatment of CRKP and hvKP infections is facing a great challenge with high mortality.29 The current study analyzes the differences between resistance genes and virulence genes through NGS concerning eleven CRKP and one hvKP isolated from aseptic body fluid. Based on the tigecycline combinations susceptibility testing in vitro, we can inform the effective choice of clinical treatment.
In our hospital, the isolation rates of CRKP increased from 3.3% in 2017 to 5.3% in 2018. It was higher than the mean level of that in Shanxi province (1.5%) in 2018 (www.chinets.com/Chinet). In our study, CRKP was commonly isolated from patients with a history of ICU, invasive procedures, and prior to carbapenem exposure. These factors are considered as risk factors in the occurrence of CRKP in previous studies.30–32 In addition, a significant portion of strains were obtained from blood specimens, suggesting that CRKP bloodstream infection was a major problem in this study.
Carbapenems are used as a last resort to treat CRKP infections.3 Worryingly, in our study, more than 80% of CRKP patients had high resistance to carbapenems. The major cause in carbapenem-resistance was CRKP strains carrying different resistance genes.33 57.1% of all CRKP carried NDM gene, similar to a study from Northwestern China16 and different with the previous studies stating that KPC is prevalent in the US, some of Europe region and most of China.7,34 We also identified NDM-1 and NDM-5 types of metallo-β-lactamases genes. Alarmingly, NDM-1 or NDM-5 has prevalent in KP in China, Poland, Greece, and Pakistan.10,19 Thus, NDM-5 CRKP strains should be actively monitored in our region. Furthermore, SIM-producing strains were rarely reported. Two SIM-producing strains were detected in this study, and one showed a co-existence of NDM-5 resistant gene.
Class B β-lactamases exhibit the highest hydrolytic activity among carbapenemases.16 NDM carbapenemases are susceptible only to ATM.10 Our data displayed that NDM-1- and NDM-5-producing strains have different MICs for most antibiotics. This might be ascribed to NDM-5 enzyme is different from the NDM-1 enzyme due to the amino acid position.10 In the present study, NDM-5-producing CRKP was sensitive to ATM and quinolones with lower MIC values. Conversely, NDM-1-producing CRKP was resistant to them, which were agreed with another report.32 Interestingly, a study showed that NDM-5-producing KP strains have high resistance to quinolones,35 while, other studies proposed controversial points.10,13 Hence, the diversity of carbapenemases among CRKP strengthens the difficulty for infection control.3,36 The remaining strains without carbapenemase resistance genes have the same resistant rates with the strains carrying carbapenemase. This suggests that the resistance to the antibiotics might attribute to new mechanisms and further studies should be conducted.32 Furthermore, only hvKP strain was sensitive to carbapenem, it suggested that carbapenems might be an effective drug against SIM-producing hvKP strain. While this result might be due to the limited strain.
In addition to expressing varied enzymes, we found multiple resistant elements leading to multi-drug resistance and performed NGS on CTX-M-3-producing ST147 CRKP, NDM-1-producing ST11 CRKP, and SIM-producing hvKP. NDM-1-producing and CTX-M-3-producing strains both possessed genes that are responsible for resistance to aminoglycosides [AAC(3)-IIb], macrolides (mphA, Mrx) third generation-cephalosporins and aztreonam (blaCTX-M-3), and quinolones [oqxA, qnrS genes]. This funding is consistent with a previous study, which proved that NDM-1 positive strains harbored qnrS1 gene.37 Studies showed that ESBL encoding genes and plasmid-mediated quinolone resistance genes can be cotransferred with the same mobile genetic elements.24,38 Consistent with this, NDM-producing CRKP harbored blaSHV-187, oqxA, and oqxB. Besides CTX-M-3-producing strain co-harbored qnrB genes. CTX-M-3-producing strain also harbored tet (C) gene, which was related to the resistance of TGC.
Although hvKP strain was sensitive to the majority of antibiotics, it still harbored an amount of resistance genes. SIM-producing hvKP strain harbored blaSHV-71 and oqxA genes. As oqxA only can confer low-level resistance to quinolones, hvKP carrying oqxA may be susceptible to quinolones.17 Moreover, SIM-producing hvKP strain was still susceptible to cephalosporins and monobactams antibiotics in our study. Additionally, fosA2 gene was only explored in hvKP strain. Compared the resistance genes of hvKP with other two CRKP strains, we found hvKP strain harbored more efflux pumps genes. From the above, we speculated that antibiotic-resistance was attributed to multiple factors, thereby causing multi-drug resistance.
In MLST analysis, ST11 was the predominant type and the majority of them carried NDM-1 gene – different from the studies showing that KPC-producing ST11 strains are the most abundant KP in China.16,19 Moreover, we found a high level of diversity of STs, among them, ST11 and ST1764 have been reported in hvKP strains.19 Serotype is another important feature of hvKP.24 The serotype of the ST1764 hvKP was K64. KPC-2-producing CRKP strain was K47, which is the most common serotype among KPC-producing KP strains in China.39,40 The SIM-producing K64 ST1764 hvKP strain was first found in our study. K64 has emerged in a large-scale study in China.41
We screened several virulence-associated genes through PCR. Both ST11 CRKP95 and ST1764 hvKP were positive for rmpA, rmpA2, and iroN genes, whereas ST11 CRKP95 was non-hypervirulent KP (non-hvKP) (negative for the string test). This could be attributed to the rmpA2 that could frame shift and introduce an internal stop codon.42 The further studies on CRKP95 are essential. Only rmpA2 was detected in ST1764 hvKP by NGS. Consistent with the previous report showing that 55% to 100% of hvKP strains express at least one copy of rmpA or rmpA2, compared to 7% to 20% of non-hvKP strains.6 Thus, hv and HM are not necessarily related.33 hvKP strain carried a high number of siderophores. Aerobactin is the most important factor of hv. In our study, only hvKP harbored the iucABCD. Surprisingly, hvKP did not carry iutA, while the iucABCD, iutA and rmpA were found to exist on the same virulence plasmid.6 Recent studies proposed that the genes encoding HM and iron acquisition as clear markers for hvKP identification.16,18,19 However, our data showed that iutA, iroE and iroN were detected in CRKP strains, albeit the results of PCR were negative. Thus, to find more accurate gene markers and identify the hvKP is crucial and urgent.33
What is more, the hvKP strains frequently cause invasive infections, such as PLA,6,19 which are consistent with our study. In ST1764 hvKP, alls genes were found, yet not kfuABC genes. These genes were associated with PLA and could increase virulence.6,43 Other virulence factors and genomic features also contribute to the enhancement of the virulence,33 which should be focused on in the subsequent studies.
TGC has high activity against CRKP in our study. As reported, TGC combined carbapenems are often used in combination therapy against CRKP strains.43,44 ATM is considered as the effective drug for the infections caused by NDM-producing strain.34 We carried out the synergism test in vitro using TGC as the main drug. The data showed that the combinations of TGC with carbapenem were effective treatments of CRKP infections.44 TGC+IPM was more effective. Our results inform an available selection for clinical treatment.
Conclusion
NDM was the main resistance mechanism of CRKP in our hospital. NDM-5-producing ST690 CRKP strain and SIM-producing ST1764 hvKP stain were first found in our study; both strains have a reduced resistance to available antibiotics. The detailed feature of resistance and virulence of CRKP and hvKP strains requires further exploration. Moreover, TGC combined with carbapenem could serve as an available treatment for CRKP infections. Our results provide a basis for selecting the effective treatment for CRKP and hvKP infections.
Acknowledgments
We thank the Department of Pharmacy, Second Hospital of Shanxi Medical University, for supporting this research. This work was supported by the Shanxi Province Natural Science Foundation (grant number 201803D31124). The sponsor had no involvement in any of the stages from the study design to submission of the paper for publication.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Walker KA, Miner TA, Palacios M, et al. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio. 2019;10:2. doi:10.1128/mBio.00089-19
2. Lepuschitz S, Schill S, Stoeger A, et al. Whole genome sequencing reveals resemblance between ESBL-producing and carbapenem resistant Klebsiella pneumoniae isolates from Austrian rivers and clinical isolates from hospitals. Sci Total Environ. 2019;662:227–235. doi:10.1016/j.scitotenv.2019.01.179
3. Xie S, Fu S, Li M, et al. Microbiological Characteristics of Carbapenem-Resistant Enterobacteriaceae Clinical Isolates Collected from County Hospitals. Infect Drug Resist. 2020;13:1163–1169. doi:10.2147/IDR.S248147
4. Chung H, Pham KA, Thanh D, et al. A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol Med. 2015;7(3):227–239. doi:10.15252/emmm.201404767
5. Heinz E, E H, Bartholdson Scott J, et al. Resistance mechanisms and population structure of highly drug resistant Klebsiella in Pakistan during the introduction of the carbapenemase NDM-1. Sci Rep. 2019;9(1):2392. doi:10.1038/s41598-019-38943-7
6. Paczosa MK. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
7. Zhu C, Liyanapathirana V, Li C, et al. Characterizing mobilized virulence factors and multidrug resistance genes in carbapenemase-producing Klebsiella pneumoniae in a Sri Lankan Hospital. Front Microbiol. 2018;9:2044.
8. Qi Y, Wei Z, Ji S, et al. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66:307–312. doi:10.1093/jac/dkq431
9. Wei Z-Q, Du -X-X, Yu Y-S, Shen P, Chen Y-G, Li L-J. Plasmid-Mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007;51(2):763–765. doi:10.1128/AAC.01053-06
10. Kong Z, Cai R, Cheng C, et al. First reported nosocomial outbreak of NDM-5-producing Klebsiella pneumoniae in a neonatal unit in China. Infect Drug Resist. 2019;12:3557–3566.
11. Garcia-Fulgueiras V, Zapata Y, Papa-Ezdra R, et al. First characterization of K. pneumoniae ST11 clinical isolates harboring blaKPC-3 in Latin America. Revista Argentina de Microbiología. 2019. doi:10.1016/j.ram.2019.10.003
12. Liu Y, Wan LG, Deng Q, Cao XW, Yu Y, Xu QF. First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol Infect. 2014;143(2):376–384. doi:10.1017/S0950268814000995
13. Wang Z, Li M, Shen X, et al. Outbreak of blaNDM-5-Harboring Klebsiella pneumoniae ST290 in a Tertiary Hospital in China. Microb Drug Resist. 2019;25(10):1443–1448. doi:10.1089/mdr.2019.0046
14. Yang Q, Wang H, Sun H, Chen H, Xu Y, Chen M. Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: results from large hospital-based surveillance studies in China. Antimicrob Agents Chemother. 2010;54(1):573–577. doi:10.1128/AAC.01099-09
15. Li H, Zhang J, Liu Y, et al. Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn Microbiol Infect Dis. 2014;78(1):63–65. doi:10.1016/j.diagmicrobio.2013.10.002
16. Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
17. Li J, Zhang H, Ning J, et al. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob Resist Infect Control. 2019;8:44. doi:10.1186/s13756-019-0489-3
18. Moura Q, Esposito F, Fernandes MR, et al. Genome sequence analysis of a hypermucoviscous/hypervirulent and MDR CTX-M-15/K19/ST29 Klebsiella pneumoniae isolated from human infection. Pathog Dis. 2017;75:9. doi:10.1093/femspd/ftx121
19. Liu Z, Gu Y, Li X, et al. Identification and characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann Lab Med. 2019;39(2):167. doi:10.3343/alm.2019.39.2.167
20. CaLS I. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2019.
21. FaDA s. TYGACIL® (tigecycline) for injection, for intravenous use. Initial US. 2016.
22. I C. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2016.
23. Hong M-Y, Hsieh -C-C, Yang C-Y, et al. A simple scoring algorithm that predicts abscesses in adults with community-onset Klebsiella pneumoniae bacteremia: hypermucoviscosity matters. Infect Drug Resist. 2020;13:1045–1055. doi:10.2147/IDR.S240809
24. Liu Y, Du F-L, Xiang T-X, et al. High prevalence of plasmid-mediated quinolone resistance determinants among serotype K1 hypervirulent Klebsiella pneumoniae isolates in China. Microbial Drug Resist. 2019;25(5):681–689. doi:10.1089/mdr.2018.0173
25. Yu F, Lv J, Niu S, et al. Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (sequence type 258 [ST258] and ST11) and hypervirulent (ST23, ST65, ST86, and ST375) strains. J Clin Microbiol. 2018;56:9. doi:10.1128/JCM.00731-18
26. Brisse S, P V, Haugaard AB, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51(12):4073–4078. doi:10.1128/JCM.01924-13
27. Fu Y, Xu M, Liu Y, Li A, Zhou J. Virulence and genomic features of a blaCTX-M-3 > and blaCTX-M-14 coharboring hypermucoviscous Klebsiella pneumoniae of serotype K2 and ST65. Infect Drug Resist. 2019;12:145–159. doi:10.2147/IDR.S187289
28. Morici P, W F, Rizzato C, et al. Synergistic activity of synthetic N-terminal peptide of human lactoferrin in combination with various antibiotics against carbapenem-resistant Klebsiella pneumoniae strains. Eur J Clin Microbiol Infect Dis. 2017;36(10):1739–1748. doi:10.1007/s10096-017-2987-7
29. Hu F, Guo Y, Yang Y, et al. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur J Clin Microbiol Infect Dis. 2019;38(12):2275–2281. doi:10.1007/s10096-019-03673-1
30. Chang H, W J, Zhou W, et al. Risk factors and mortality for patients with Bloodstream infections of Klebsiella pneumoniae during 2014–2018: clinical impact of carbapenem resistance in a large tertiary hospital of China. J Infect Public Health. 2020;13:784–790.
31. Chen X, Liu Q, Liu W-E, Yan Q. Risk factors for subsequential carbapenem-resistant Klebsiella pneumoniae clinical infection among rectal carriers with carbapenem-resistant Klebsiella pneumoniae. Infect Drug Resist. 2020;13:1299–1305. doi:10.2147/IDR.S247101
32. Pan F, Tian D, Wang B, et al. Fecal carriage and molecular epidemiology of carbapenem-resistant Enterobacteriaceae from outpatient children in Shanghai. BMC Infect Dis. 2019;19(1):678. doi:10.1186/s12879-019-4298-3
33. Zhao Y, Zhang S, Fang R, et al. Dynamic epidemiology and virulence characteristics of carbapenem-resistant Klebsiella pneumoniae in Wenzhou, China from 2003 to 2016. Infect Drug Resist. 2020;13:931–940. doi:10.2147/IDR.S243032
34. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi:10.1093/femsre/fux013
35. Muhammad U, Qamar TRW, Toleman MA, et al. Dissemination of genetically diverse NDM-1, −5, −7 producing-Gram-negative pathogens isolated from pediatric patients in Pakistan. Future Microbiol. 2019;14:691–704. doi:10.2217/fmb-2019-0012
36. Alizadeh N, Ahangarzadeh Rezaee M, Samadi Kafil H, et al. Evaluation of resistance mechanisms in carbapenem-resistant Enterobacteriaceae. Infect Drug Resist. 2020;13:1377–1385. doi:10.2147/IDR.S244357
37. Mukherjee S, Bhattacharjee A, Naha S, et al. Molecular characterization of NDM-1-producing Klebsiella pneumoniae ST29, ST347, ST1224, and ST2558 causing sepsis in neonates in a tertiary care hospital of North-East India. Infect Genetics Evol. 2019;69:166–175. doi:10.1016/j.meegid.2019.01.024
38. Higashino M, Murata M, Morinaga Y, et al. Fluoroquinolone resistance in extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a Japanese tertiary hospital: silent shifting to CTX-M-15-producing K. pneumoniae. J Med Microbiol. 2017;66(10):1476–1482. doi:10.1099/jmm.0.000577
39. Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. doi:10.1093/jac/dkz446
40. Huang Y-H, Chou S-H, Liang S-W, et al. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73(8):2039–2046. doi:10.1093/jac/dky164
41. De Campos TA, Gonçalves LF, Magalhaes KG, et al. A fatal bacteremia caused by hypermucousviscous KPC-2 producing extensively drug-resistant K64-ST11 Klebsiella pneumoniae in Brazil. Front Med (Lausanne). 2018;5:265. doi:10.3389/fmed.2018.00265
42. Lu Y, Feng Y, McNally A, Zong Z. Occurrence of colistin-resistant hypervirulent Klebsiella variicola. J Antimicrob Chemother. 2018;73(11):3001–3004. doi:10.1093/jac/dky301
43. Roe CC, Vazquez AJ, Esposito EP, Zarrilli R, Sahl JW. Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front Microbiol. 2019;10:542.
44. Zhang J, Yu L, Fu Y, et al. Tigecycline in combination with other antibiotics against clinical isolates of carbapenem-resistant Klebsiella pneumoniae in vitro. Ann Palliat Med. 2019;8(5):622–631. doi:10.21037/apm.2019.09.11
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.