Back to Journals » Infection and Drug Resistance » Volume 12
Emergence of multidrug resistance and extensive drug resistance among enterococcal clinical isolates in Egypt
Authors Said HS , Abdelmegeed ES
Received 1 October 2018
Accepted for publication 18 January 2019
Published 7 May 2019 Volume 2019:12 Pages 1113—1125
DOI https://doi.org/10.2147/IDR.S189341
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Heba Shehta Said, Eman Salama Abdelmegeed
Department of Microbiology and Immunology, Faculty of Pharmacy, Mansoura University, Mansoura 35516, Egypt
Introduction: Enterococci commonly inhabit the gastrointestinal tract of both human and animals; however, they have emerged as a leading cause of several infections with substantial morbidity and mortality. Their ability to acquire resistance combined with intrinsic resistance to various antimicrobials makes treatment of enterococcal infections challenging.
Materials and methods: The aim of the study was to evaluate the antimicrobial resistance pattern, and assess the prevalence of multidrug resistance (MDR) and extensive drug resistance (XDR) among enterococcal isolates, collected from different clinical sources, in Mansoura University Hospitals, Egypt.
Results: Antibiotic sensitivity testing revealed elevated levels of resistance among enterococcal clinical isolates (N=103). All E. faecium (N=32) and 74.6% of E. faecalis isolates (N=71) were MDR, while two E. faecalis and four E. faecium isolates were XDR. High level gentamicin resistance was detected in 79.6%, most of them carried the aac(6’)-Ie-aph(2’’)-Ia gene. High level streptomycin resistance was seen in 36.9%, of which 52.6% carried the ant(6’)-Ia gene. Resistance to macrolides and lincosamides were mediated by ermB (92.2%) and msrA/B (42.7%). tetK, tetL, and tetM genes were detected among tetracyclines resistant isolates. Resistance to vancomycin was detected in 15.5%, where vanB and vanC1 gene clusters were detected in VRE isolates. Ten isolates (9.7%) were resistant to linezolid, eight of which harbored the optrA gene. Vancomycin and linezolid resistant enterococci were more likely to exhibit strong/moderate biofilm formation than vancomycin and linezolid sensitive ones.
Conclusion: Elevated levels of resistance to different classes of antimicrobial agents and emergence of MDR and XDR strains pose a major threat with limited therapeutic options for infections caused by this emerging pathogen.
Keywords: enterococci, antibiotic resistance, biofilm formation, RAPD typing, MDR enterococci, XDR enterococci, VRE, linezolid resistant enterococci
Introduction
Enterococci are Gram-positive, facultative-anaerobic cocci that are persistent in nature and can survive under adverse environmental conditions for prolonged periods of time.1 They are considered commensal organisms and members of the healthy intestinal microbiota of humans and animals.1,2 However, in the last two decades, they have emerged as one of the leading causes of several hospital-acquired and community-acquired infections with substantial morbidity and mortality.3 The most common infections caused by enterococci are urinary tract infections (UTIs), bacteremia, peritonitis, cholecystitis, meningitis, wound infections, endocarditis, and neonatal infections.3 Enterococci are considered the second etiologic agent of UTIs and the third of nosocomial bacteremia.1–3 Although E. faecalis is not considered as a member of the healthy oral microbiota, it has been reported in common dental diseases as dental caries and peridontitis.4–6
E. faecalis and E. faecium are the most commonly reported enterococcal species, counting for up to 90% of enterococcal infections, while other enterococcal species are rarely reported to cause human infections.2,7 Being equipped with an array of virulence determinants and capability to survive in hospital setting makes infections difficult to manage.1,2 Antimicrobial therapy of infections caused by enterococci is problematic because of its intrinsic reduced susceptibility to several frequently used antimicrobial agents including: aminoglycosides (except for high level resistance), clindamycin, cephalosporins, and trimethoprim/sulfamethoxazole.8,9 Moreover, acquired resistance through lateral gene transfer to other available antimicrobial agents, including: beta-lactams, aminoglycosides (high level resistance), macrolides, glycopeptides, and oxazolidinones, makes it more challenging.10 Vancomycin resistant enterococci (VRE) have been reported as a leading cause of outbreaks of hospital-acquired infections and in ICU-hospitalized patients.11,12 Linezolid was the drug of choice for management of infections caused by VRE; however, linezolid resistant enterococci have emerged and are reported.13,14
Clinicians may face very limited treatment options as a result of selection and spread of multidrug-resistant (MDR) and extensively drug resistant (XDR) strains.10,15 Magiorakos et al16 have defined MDR as non-susceptibility to at least one agent in three or more antimicrobial classes, while XDR has been defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial classes. Recently, enterococci have been ascribed in biofilm-associated infections of medical devices.17 The ability of enterococci to form a biofilm amplifies the difficulty of treatment due to increased antimicrobial resistance in the biofilm.18–20 Therefore, antimicrobial susceptibility testing to all available antimicrobial agents is exceptionally elemental for deciding the proper treatment of enterococcal infections.
The aim of this study is to evaluate the antimicrobial resistance pattern and assess the prevalence of MDR and XDR among enterococcal isolates collected from different clinical sources in Mansoura University Hospitals, Egypt. Moreover, the study aims to explore the molecular basis of resistance to different classes of antimicrobial agents.
Materials and methods
Bacterial isolates
Enterococcal isolates (N=103) were recovered from different clinical sources in Mansoura university Hospitals, Egypt between January and August 2017. Identification of enterococcal isolates was based on standard biochemical tests,21,22 then confirmed by a multiplex PCR system.23,24
Antimicrobial susceptibility testing
The antimicrobial susceptibility profile of enterococcal isolates was determined using a disc diffusion method.25 The inhibition zones were interpreted according to the recommendations of the Clinical and Laboratory Standard Institute.26 Susceptibility to different classes of antimicrobial agents was tested, including: ampicillin (10 µg), ampicillin/sulbactam (20 µg/10 µg), Amoxicillin (20 µg), amoxicillin/clavulanic acid (20 µg/10 µg), Imipenem (10 µg), Meropenem (10 µg), vancomycin (30 µg), clindamycin (2 µg), erythromycin (15 µg), clarithromycin (15 µg), azithromycin (15 µg), ciprofloxacin (5 µg), Levofloxacin (5 µg), tertracycline (30 µg), doxycycline (30 µg), and Linezolid (30 µg) (Oxoid, Basingstoke, UK).
The minimum inhibitory concentration (MIC) for vancomycin and linezolid was also determined by standard broth microdilution method, as per Clinical & Laboratory Standards Institute (CLSI) guidelines.26 High-level gentamicin resistance (HLGR, 500 µg/mL) and high-level streptomycin resistance (HLSR, 1000 µg/mL) were determined according to CLSI guidelines.26
Biofilm assay
A quantitative assay of biofilm formation capacity of enterococcal isolates was carried out using a polystyrene microtiter plate assay, as described previously.27,28
Molecular assays
Multiplex PCR analysis of vancomycin-resistant enterococcus species
Total genomic DNA was prepared as described previously.29 Multiplex PCR was used for the detection of vancomycin resistance determinants, including: vanA, vanB, vanC1, and vanC2/3 genes, besides primers for the detection of the ddl genes of E. faecium and E. faecalis and of 16S rRNA as a PCR control (Table 1), as described previously.23,24
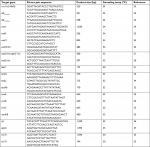  | Table 1 List of primers used in this study |
Detection of antimicrobial resistance encoding genes by PCR
The prevalence of resistance determinants to different classes of antimicrobial agents among enterococcal clinical isolates was examined. Genes encoding aminoglycosides,30,31 tetracyclines,32–34 macrolide,35 vancomycin,23,24 and linezolid36,37 resistance were detected by PCR. Primer pairs used and expected amplicon sizes are listed in Table 1.
Genotyping by random amplified polymorphic DNA (RAPD-PCR) analysis
All enterococcal isolates were genotyped by RAPD-PCR, as described previously.38,39 RAPD patterns were analyzed using gelJ software.40 A similarity matrix was estimated using Dice’s coefficient, and a dendrogram was created based on the unweighted-pair group method with arithmetic averages (UPGMA).41 Enterococcal isolates with a similarity coefficient ≥85% were considered as the same genotype/clonal.
Statistical analysis
Correlation between the capacity of enterococcal clinical isolates for biofilm formation, clinical source of collection, and resistance to different classes of antimicrobial agents were evaluated using Fisher’s exact test or chi-squared test, where a P-value<0.05 was considered statistically significant. Data were analyzed using the SPSS software (version 20.0; SPSS, Chicago, IL, USA).
Results
During the period of study, 103 enterococcal isolates were collected from Mansoura University Hospitals, Egypt from different clinical sources including: urine (56 samples, 54.4%), blood (36 samples, 35%), and wounds (11 samples, 10.7%). All isolates were identified as enterococci phenotypically and genotypically. Seventy-one isolates (68.9%) were identified as E. fecalis, while 32 isolates (31.1%) were identified as E. faecium.
Antimicrobials susceptibility of enterococcal isolates
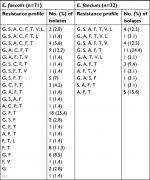
Among the tested enterococcal isolates, resistance to β-lactam antibiotics was distributed as follows: ampicillin (51.5% R), ampicillin/sulbactam (50.5% R, 1% I), amoxicillin and amoxicillin/clavulanic acid (47.6% R, 3.9% I). There was significant difference in resistance to β-lactam antibiotics between different enterococcal species (P<0.001) (Table S1). All E. faecium isolates (N=32) were resistant β-lactam and β-lactam/β-lactamase inhibitor combinations, while only 29.6% of E. faecalis isolates (N=71) were resistant (Table 2). blaZ gene was not detected in any enterococcal isolate. E. faecalis resistance to imipenem was (21.1% R), and meropenem was (26.8% R, 4.2% I).
Screening of high-level resistance to aminoglycosides has indicated that 82 isolates (79.6%) showed HLGR, while 38 isolates (36.9%) showed HLSR (Table 2). Among the isolates that showed HLGR, 75 isolates (91.5%) carried the aac(6’)-Ie-aph(2’’)-Ia gene (Figure S1). Among the isolates that showed HLSR, 20 isolates (52.6%) carried the ant(6’)-Ia gene. No significant difference was observed in prevalence of HLGR between both enterococcal species; however, E. faecium isolates showed a higher rate of HLSR than E. faecalis isolates (P<0.001) (Table S1).
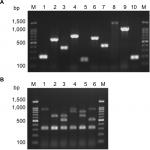
Resistance to macrolides was distributed as follows: erythromycin (91.3% R, 8.7% I), azithromycin (74.6% R, 1.4% I), and clarithromycin (85.9% R, 0% I). All isolates were resistant to clindamycin (Table 2). Among the genes encoding macrolide resistance, ermB was detected in 95 isolates (92.2%), while msrA/B was detected in 44 isolates (42.7%) (Figure S1). Both ermB and msrA/B genes were detected in 40 isolates (38.8%). ermA and ermC were not detected in any enterococcal isolate (Figures 1 and 2).
Resistance to tetracycline was (93.2% R, 5.8% I), while resistance to doxycycline was (55.3% R, 31.1% I) (Table 2). E. faecium isolates showed a higher rate of resistance to doxycycline than E. faecalis isolates (P=0.016) (Table S1). The tetK gene was detected in 62 isolates (60.2%), the tetL gene in 71 isolates (68.9%), and tetM in 99 isolates (96.1%) (Figure S1). tetK, tetL, and tetM were detected in 37 isolates (35.9%), while two out of the three tested genes were detected in 55 isolates (53.4%) (Figures 1 and 2).
Determination of MIC by broth microdilution method has indicated that seven isolates (6.8%) were resistant to vancomycin (MIC≥32 µg/mL), while nine isolates (8.7%) were intermediately resistant (MIC=8–16 µg/mL) (Table 2). A higher rate of vancomycin resistance was observed among E. faecium isolates compared to E. faecalis (P<0.001) (Table S1). Multiplex PCR analysis of vancomycin resistant enterococci has indicated that six isolates carried the vanB gene cluster (MIC=8–>256 µg/mL), while 10 isolates carried vanC1 (MIC=8–32 µg/mL) (Figure S1). VanA and vanC2/3 gene clusters were not detected in any isolate (Figures 1 and 2).
Ten isolates (9.7%) were resistant to linezolid (MIC≥8 µg/mL). Among linezolid resistant strains, eight isolates carried the optrA gene (Figure S1), while the cfr gene was not detected in any isolate. Prevalence of quinolone resistance among enterococcal isolates was (81.6% R, 3.9% I) for ciprofloxacin and (89.3% R, 2% I) for levofloxacin (Table 2).
Biofilm formation among enterococcal isolates
The capacity of enterococci isolated from different clinical sources to form a biofilm was evaluated by polystyrene microtiter plate assay, as previously described, and divided into strong, moderate, weak biofilm producers, and non-biofilm producers.27,28 The ability to form a biofilm was detected in almost all clinical isolates examined (97 isolates, 94.2%). Only six isolates (5.8%) were biofilm non-producers (Table S2). Among the biofilm forming isolates, 21 isolates (20.4%) were strong biofilm producers, 41 isolates (39.8%) were moderate biofilm producers, while 35 isolates (34%) were classified as weak biofilm producers. No significant difference in biofilm formation capacity was observed between E. faecalis and E. faecium isolates (P>0.05) (Table S2). Biofilm formation capacity was similar between different clinical sources from which the isolates were collected (P>0.05).
Correlation between biofilm formation capacity and resistance to different classes of antimicrobials has indicated that vancomycin-resistant isolates were more likely to exhibit a strong or moderate biofilm formation capacity than vancomycin-sensitive isolates (P=0.034) (Table S3). Moreover, all linezolid-resistant strains exhibited strong or moderate biofilm formation capacity (P=0.032). Resistance to β-lactam antibiotics and β-lactam/β-lactamase combinations significantly correlated with biofilm formation capacity among enterococcal clinical isolates (P=0.039) (Table S3). However, there was no significant correlation between biofilm formation ability and resistance/sensitivity to other classes of antimicrobial agents.
Clonal relationship analysis by RAPD-PCR
Enterococcal isolates recovered from different clinical sources were genotyped by the RAPD-PCR approach. Great diversity was observed in RAPD-PCR profiles of both E. faecalis (Figure 1) and E. faecium (Figure 2) isolates recovered from different clinical sources at an 85% cutoff. Some E. faecalis isolates showed a similarity coefficient higher than 85% and were considered clonal (for example: E.fs.43 and E.fs.42), however, their antibiotic susceptibility profiles and resistance determinants detected were different (Figure 1). No clustering of the enterococcal isolates was observed based on the clinical source from which they were recovered or their biofilm formation capacity.
Discussion
Enterococci colonize the gastrointestinal tract of both humans and animals and are considered a part of its normal beneficial flora;1 however, it has emerged as a nosocomial pathogen in the last few decades.2 E. faecalis is the most commonly reported species accounting for more than 80% of enterococcal infections; however, E. faecium has also been increasingly reported which could be attributed to its resistance to multiple classes of antibiotics.1,2 In our study, E. faecalis was the most prevalent species detected, accounting for 68.9%, while 31.1% were identified as E. faecium. Most of the enterococcal isolates (54.4%) were collected from urine, followed by blood (35%), and wound (10.7%). Previous reports have also indicated that UTI is the most common infection caused by enterococci, followed by surgical site infections (SSI), bacteremia, and endocarditis.42–44
Recent reports have indicated that HLGR among enterococcal isolates ranged from 50% to 65%, while HLSR was less common than HLGR, ranging from 29% to 38%.43,45,46 Similarly, we had observed HLGR to be more predominant than HLSR. However, our study has indicated a higher rate of HLGR (79.6%) among enterococcal isolates in Egypt. Although enterococci have intrinsic resistance to low levels of aminoglycosides, high-level resistance is mediated by acquisition of aminoglycoside-modifying enzymes (AMEs) encoding genes. HLGR (MIC≥500 mg/mL) in enterococci is mostly mediated by the aac(6’)-Ie-aph(2’’)-Ia gene that encodes the bifunctional aminoglycoside modifying enzyme AAC(6’)-APH(2’’) and confers high-level resistance to aminoglycosides except streptomycin, while HLSR in enterococci are mediated by ant(6’)-Ia gene. In enterococci, AMEs eliminate the synergistic activity of aminoglycosides when combined with a cell wall active agent, such as ampicillin or vancomycin.43,45,46
Increased resistance to macrolides, lincosamides, and streptogramin B (MLSB resistance) among enterococcal clinical isolates may be attributed to the wide use of this class of antimicrobials for treatment of enterococcal infections, except UTIs.47 MLSB resistance is attributed to active efflux, target modification, or inactivating enzymes.48 Methylation of 23S rRNA, mediated by erm gene, mediates resistance to macrolides, lincosamides, and streptogramin B. ermB is the most commonly reported gene among enterococcal isolates, while ermA and ermC are occasionally reported.49 msrA/B gene encodes efflux pump for macrolides and streptogramin B.47 Similar to previous reports, ermB and msrA/B genes were present in most macrolide resistant enterococcal isolates tested.45,47,49
Tetracyclines are broad-spectrum antibiotics and one of the most frequently used antimicrobials in Europe and in veterinary medicine.47 Tetracycline resistance determinants (tet genes) mediate ribosomal protection, enzymatic inactivation, and efflux of tetracyclines. tetK and tetL are involved in the efflux mechanism, while tetM plays a role in ribosomal protection among enterococci.50 Our data have indicated that most enterococcal clinical isolates (89.3%) carried more than one tet gene conferring different mechanisms of resistance to tetracyclines (efflux and ribosomal protection). There is not enough data or reports describing the prevalence of tetracycline resistance and its determinants among enterococcal clinical isolates. Recently, Zahid et al51 indicated that 57% of enterococcal isolates carried tetL, 50% carried tetM, while 33% carried both tetL and tetM.
Association of antibiotic resistance genes with mobile genetic elements increases the risk of dissemination of antibiotics resistance among enterococcal isolates. Tn916 was the first conjugative transposon, carrying the tetM gene, identified in E. faecalis.52 Since then, a large number of transposable elements have been identified carrying resistance to different classes of antibiotics.47 In the present study, the integrase (int) and the excisase (xis) genes of Tn916 were detected in 72 isolates (69.9%) of enetrococcal isolates collected from different clinical sources. A recent study has indicated that Tn916 was detected in 43.3% of enterococcal clinical isolates.51
Enterococcal resistance to glycopeptides is mediated by van operons.53 Most vancomycin resistant enterococcal outbreaks are attributed to vanA and vanB gene clusters.43,54 vanA is inducible and confers high-level resistance to both vancomycin and teicoplanin, while vanB confers a variable level of resistance to vancomycin, but not to teicoplanin.53 vanC is less virulent than vanA and vanB and intrinsically present in some enterococcal species. However, the vanC1 gene cluster has been recently detected in E. faecalis and E. faecium clinical isolates providing moderate level resistance to vancomycin.55,56 Variable rates of vancomycin resistance were detected across the world, with lower rates detected in Europe and Canada (4%–6%).54,57,58 A recent study in Iran has indicated that 18.8% of their enterococcal clinical isolates were resistant to vancomycin, where all isolates had MIC>256% and 90% of VRE carried the vanA gene cluster.43 Our data showed high prevalence of VRE in Egypt (15.5%); however, VRE carried either vanB or vanC1 gene clusters. Moreover, our results coincide with previous reports that showed a higher rate of vancomycin resistance in E. faecium than E. faecalis clinical isolates.54
Linezolid, the first oxazolidinone used clinically, is one of the most promising agents against MDR G+ve pathogens, especially VRE. However, linezolid resistance has been increasingly reported among enterococcal clinical isolates.59,60 Klare et al59 found that the rate of linezolid resistance increased in Germany from <1% in 2008 to >9% in 2014. Our results coincide with their results, where 9.7% of our isolates were linezolid resistant. Mutation in domain V of the 23S rRNA gene, and transferable plasmid mediated methytransferase (cfr) and optrA genes have been reported to mediate resistance to oxazolidinones.36,37 Beside oxazolidinones, the optrA gene also confers resistance to phenolics. It was first described in 2015 in enterococci recovered from human and animal origins in China.37 Afterwards, the optrA gene has been detected in linezolid resistant enterococcal isolates in several places worldwide.61–63 In our study, the optrA gene was detected in 80% of the linezolid resistant enterococci. To our knowledge, this is the first report of the optrA gene among linezolid resistant enterococci in Egypt. The cfr gene has been previously detected among linezolid resistant enterococci;36,59,60 however, it was not detected among linezolid resistant enterococci tested.
Enterococci have a high biofilm formation capacity, which increases the difficulty of management of enterococcal infections and hinders the penetration of antibiotics.64,65 In the present study, most of the enterococcal clinical isolates were biofilm producers, where 60.2% were strong/moderate biofilm producers. Several previous studies have indicated a high biofilm formation capacity among enterococcal clinical isolates.64,65 All linezolid-resistant strains exhibited strong/moderate biofilm formation. Moreover, vancomycin-resistant isolates were more likely to exhibit strong/moderate biofilm formation than vancomycin-sensitive isolates. A previous study has also indicated the correlation between biofilm formation capacity among enterococcal clinical isolates and resistance to linezolid.65 Another study has indicated that resistance to penicillin G, ampicillin, vancomycin, ciprofloxacin, chloramphenicol, and nitrofurantoin was significantly higher among biofilm producers than biofilm non-producers.66
In this study, a higher level of resistance was observed among E. faecium than E. faecalis isolates (Table 2) as previously reported.2,43,45,59 Magiorakos et al16 have defined MDR as non-susceptibility to at least one agent in three or more antimicrobial classes, while XDR was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial classes. According to this definition, 53 E. faecalis isolates (74.6%) were MDR, while all E. faecium (N=32) were MDR strains. Two E. faecalis isolates (2.8%) and four E. faecium (12.5%) were resistant to all tested antimicrobials tested and considered as XDR (Table S4). Horizontal gene transfer is held responsible for the elevated levels of resistance to different antimicrobials, since plasmids and transposable elements usually carry resistance determinants for more than one class of antimicrobial agents. The increased level of resistance to vancomycin and linezolid highlights the problem of limited options to treatment infections caused by MDR and XDR enterococcal strains.
Conclusion
Antimicrobial susceptibility testing is of utmost importance in formulating an effective antibiotic policy for treatment of enterococcal infections and reducing morbidity and mortality. Neglecting standard protocols, empirical use of antibiotics, and lack of sufficient information are the leading causes for the emergence and spread of MDR and XDR enterococci in hospitals. Hence, effective measures, standard protocols, and programs to control the spread of enterococci should be implemented.
Disclosure
The authors report no conflicts of interest in this work.
References
Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155(Pt 6):1749–1757. | ||
Van Tyne D, Gilmore MS. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol. 2014;68:337–356. | ||
Simmons BP, Larson EL. Multiple drug resistant organisms in healthcare: the failure of contact precautions. J Infect Prev. 2015;16(4):178–181. | ||
Dahlén G, Blomqvist S, Almståhl A, Carlén A. Virulence factors and antibiotic susceptibility in enterococci isolated from oral mucosal and deep infections. J Oral Microbiol. 2012;4(1):10855. | ||
Rams TE, Degener JE, van Winkelhoff AJ. Prevalence of β-lactamase-producing bacteria in human periodontitis. J Periodontal Res. 2013;48(4):493–499. | ||
Rams TE, Feik D, Young V, Hammond BF, Slots J. Enterococci in human periodontitis. Oral Microbiol Immunol. 1992;7(4):249–252. | ||
Hammerum AM. Enterococci of animal origin and their significance for public health. Clin Microbiol Infect. 2012;18(7):619–625. | ||
Huycke MM, Sahm DF, Gilmore MS. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4(2):239–249. | ||
Kaye KS, Kaye D. Multidrug-resistant pathogens: mechanisms of resistance and epidemiology. Curr Infect Dis Rep. 2000;2(5):391–398. | ||
Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10(4):266–278. | ||
Flokas ME, Karageorgos SA, Detsis M, Alevizakos M, Mylonakis E. Vancomycin-resistant enterococci colonisation, risk factors and risk for infection among hospitalised paediatric patients: a systematic review and meta-analysis. Int J Antimicrob Agents. 2017;49(5):565–572. | ||
Moosavian M, Ghadri H, Samli Z. Molecular detection of vanA and vanB genes among vancomycin-resistant enterococci in ICU-hospitalized patients in Ahvaz in southwest of Iran. Infect Drug Resist. 2018;11:2269–2275. | ||
Chen M, Pan H, Lou Y, et al. Epidemiological characteristics and genetic structure of linezolid-resistant Enterococcus faecalis. Infect Drug Resist. 2018;11:2397–2409. | ||
Gawryszewska I, Żabicka D, Hryniewicz W, Sadowy E. Linezolid-resistant enterococci in Polish hospitals: species, clonality and determinants of linezolid resistance. Eur J Clin Microbiol Infect Dis. 2017;36(7):1279–1286. | ||
van Harten RM, Willems RJL, Martin NI, Hendrickx APA. Multidrug-resistant enterococcal infections: new compounds, novel antimicrobial therapies? Trends Microbiol. 2017;25(6):467–479. | ||
Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. | ||
Paganelli FL, Willems RJ, Leavis HL. Optimizing future treatment of enterococcal infections: attacking the biofilm? Trends Microbiol. 2012;20(1):40–49. | ||
Parker RE, Laut C, Gaddy JA, Zadoks RN, Davies HD, Manning SD. Association between genotypic diversity and biofilm production in group B Streptococcus. BMC Microbiol. 2016;16(1):86. | ||
Sandoe JA, Witherden IR, Cove JH, Heritage J, Wilcox MH. Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J Med Microbiol. 2003;52(Pt 7):547–550. | ||
Seno Y, Kariyama R, Mitsuhata R, Monden K, Kumon H. Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med Okayama. 2005;59(3):79–87. | ||
Facklam RR, Maria da Gloria SC, Teixeira LM. History, Taxonomy, Biochemical Characteristics, and Antibiotic Susceptibility Testing of Enterococci. Washington, DC: ASM Press; 2002. | ||
Mahon CR, Lehman DC, Manuselis G. Selected procedures. Textbook of Diagnostic Microbiology. 4th ed. In: Mahon CR, Lehman D, Manuselis G, editors; Oxford: Elsevier Health Sciences; 2011:1018–1024. | ||
Elsayed S, Hamilton N, Boyd D, Mulvey M. Improved primer design for multiplex PCR analysis of vancomycin-resistant Enterococcus spp. J Clin Microbiol. 2001;39(6):2367–2368. | ||
Kariyama R, Mitsuhata R, Chow JW, Clewell DB, Kumon H. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 2000;38(8):3092–3095. | ||
Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. | ||
CLSI. Performance Standards for Antimicrobial Susceptibility Testing: 27th ed. CLSI document M100-S27 (ISBN 1-56238-805-3). 2017. | ||
Mohamed JA, Huang W, Nallapareddy SR, Teng F, Murray BE. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72(6):3658–3663. | ||
Toledo-Arana A, Valle J, Solano C, et al. The enterococcal surface protein, esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67(10):4538–4545. | ||
Englen MD, Kelley LC. A rapid DNA isolation procedure for the identification of Campylobacter jejuni by the polymerase chain reaction. Lett Appl Microbiol. 2000;31(6):421–426. | ||
Clark NC, Olsvik O, Swenson JM, Spiegel CA, Tenover FC. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob Agents Chemother. 1999;43(1):157–160. | ||
Van de Klundert J. PCR detection of genes coding for aminoglycoside-modifying enzymes. Diagn Mol Microbiol. 1993:547–552. | ||
Aminov RI, Garrigues-Jeanjean N, Mackie RI. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol. 2001;67(1):22–32. | ||
Cochetti I, Tili E, Vecchi M, et al. New Tn916-related elements causing erm(B)-mediated erythromycin resistance in tetracycline-susceptible pneumococci. J Antimicrob Chemother. 2007;60(1):127–131. | ||
Gevers D, Danielsen M, Huys G, Swings J. Molecular characterization of tet (M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl Environ Microbiol. 2003;69(2):1270–1275. | ||
Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40(11):2562–2566. | ||
Diaz L, Kiratisin P, Mendes RE, Panesso D, Singh KV, Arias CA. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob Agents Chemother. 2012;56(7):3917–3922. | ||
Wang Y, Lv Y, Cai J, et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother. 2015;70(8):2182–2190. | ||
Martín-Platero AM, Valdivia E, Maqueda M, Martínez-Bueno M. Characterization and safety evaluation of enterococci isolated from Spanish goats’ milk cheeses. Int J Food Microbiol. 2009;132(1):24–32. | ||
Muñoz-Atienza E, Araújo C, Campo Rdel, Hernández PE, Herranz C, Cintas LM. Safety assessment and molecular genetic profiling by pulsed-field gel electrophoresis (PFGE) and PCR-based techniques of Enterococcus faecium strains of food origin. LWT Food Sci Technol. 2016;65:357–362. | ||
Heras J, Domínguez C, Mata E, et al. GelJ-a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics. 2015;16(1):270. | ||
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. | ||
Arshadi M, Douraghi M, Shokoohizadeh L, Moosavian SM, Pourmand MR. High prevalence of diverse vancomycin resistance Enterococcus faecium isolates in clinical and environmental sources in ICU wards in Southwest of Iran. Microb Pathog. 2017;111:212–217. | ||
Jahansepas A, Aghazadeh M, Rezaee MA, et al. Occurrence of Enterococcus faecalis and Enterococcus faecium in Various Clinical Infections: Detection of Their Drug Resistance and Virulence Determinants. Microb Drug Resist. 2018;24(1):76–82. | ||
Somily AM, Al-Mohizea MM, Absar MM, et al. Molecular epidemiology of vancomycin resistant enterococci in a tertiary care hospital in Saudi Arabia. Microb Pathog. 2016;97:79–83. | ||
Heidari H, Emaneini M, Dabiri H, Jabalameli F. Virulence factors, antimicrobial resistance pattern and molecular analysis of enterococcal strains isolated from burn patients. Microb Pathog. 2016;90:93–97. | ||
Shete V, Grover N, Kumar M. Analysis of aminoglycoside modifying enzyme genes responsible for high-level aminoglycoside resistance among enterococcal isolates. J Pathog. 2017;2017:3256952. | ||
Marosevic D, Kaevska M, Jaglic Z. Resistance to the tetracyclines and macrolide-lincosamide-streptogramin group of antibiotics and its genetic linkage- a review. Ann Agric Environ Med. 2017;24(2):338–344. | ||
Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43(12):2823–2830. | ||
Iweriebor BC, Obi LC, Okoh AI. Macrolide, glycopeptide resistance and virulence genes in Enterococcus species isolates from dairy cattle. J Med Microbiol. 2016;65(7):641–648. | ||
Roberts MC. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19(1):1–24. | ||
Zahid S, Bin-Asif H, Hasan KA, Rehman M, Ali SA. Prevalence and genetic profiling of tetracycline resistance (Tet-R) genes and transposable element (Tn916) in environmental Enterococcus species. Microb Pathog. 2017;111:252–261. | ||
Franke AE, Clewell DB. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145(1):494–502. | ||
Ahmed MO, Baptiste KE. Vancomycin-resistant enterococci: a review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb Drug Resist. 2018;24(5):590–606. | ||
O’Driscoll T, Crank CW. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist. 2015;8:217–230. | ||
de Garnica ML, Valdezate S, Gonzalo C, Saez-Nieto JA. Presence of the vanC1 gene in a vancomycin-resistant Enterococcus faecalis strain isolated from ewe bulk tank milk. J Med Microbiol. 2013;62(Pt 3):494–495. | ||
Sun M, Wang Y, Chen Z, Zhu X, Tian L, Sun Z. The first report of the vanC1 gene in Enterococcus faecium isolated from a human clinical specimen. Mem Inst Oswaldo Cruz. 2014;109(6):712–715. | ||
Sievert D, Ricks P, Edwards J, et al; National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34(1):1–14. | ||
Zhanel GG, Adam HJ, Baxter MR, et al; Canadian Antimicrobial Resistance Alliance Antimicrobial susceptibility of 22746 pathogens from Canadian hospitals: results of the CANWARD 2007-11 study. J Antimicrob Chemother. 2013;68(Suppl 1):i7–i22. | ||
Klare I, Fleige C, Geringer U, et al. Increased frequency of linezolid resistance among clinical Enterococcus faecium isolates from German hospital patients. J Glob Antimicrob Resist. 2015;3(2):128–131. | ||
Li B, Ma CL, Yu X, et al. Investigation of mechanisms and molecular epidemiology of linezolid nonsusceptible Enterococcus faecalis isolated from a teaching hospital in China. J Microbiol Immunol Infect. 2016;49(4):595–599. | ||
Cai J, Wang Y, Schwarz S, et al. High detection rate of the oxazolidinone resistance gene optrA in Enterococcus faecalis isolated from a Chinese anorectal surgery ward. Int J Antimicrob Agents. 2016;48(6):757–759. | ||
Cavaco LM, Bernal JF, Zankari E, et al. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J Antimicrob Chemother. 2016;72(3):678–683. | ||
Vorobieva V, Roer L, Justesen US, et al. Detection of the optrA gene in a clinical ST16 Enterococcus faecalis isolate in Denmark. J Glob Antimicrob Resist. 2017;10:12–13. | ||
Anderson AC, Jonas D, Huber I, et al. Enterococcus faecalis from food, clinical specimens, and oral sites: prevalence of virulence factors in association with biofilm formation. Front Microbiol. 2015;6:1534. | ||
Zheng JX, Bai B, Lin ZW, et al. Characterization of biofilm formation by Enterococcus faecalis isolates derived from urinary tract infections in China. J Med Microbiol. Epub 2017 Nov 17. | ||
Fallah F, Yousefi M, Pourmand MR, Hashemi A, Nazari Alam A, Afshar D. Phenotypic and genotypic study of biofilm formation in enterococci isolated from urinary tract infections. Microb Pathog. 2017;108:85–90. |
Supplementary materials
  | Table S2 Prevalence of biofilm formation among enterococcal isolates collected from different clinical sources Note: aE. faecalis vs E. faecium; P-value<0.05 was considered statistically significant. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.