Back to Journals » Infection and Drug Resistance » Volume 13
Emergence of Hypervirulent Ceftazidime/Avibactam-Resistant Klebsiella pneumoniae Isolates in a Chinese Tertiary Hospital
Authors Li D , Liao W , Huang H, Du F, Wei D, Mei Y, Long D, Wan L, Liu Y, Zhang W
Received 8 April 2020
Accepted for publication 16 July 2020
Published 3 August 2020 Volume 2020:13 Pages 2673—2680
DOI https://doi.org/10.2147/IDR.S257477
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Dan Li,1,2,* Wenjian Liao,1,2,* Hai-hua Huang,3 Fang-ling Du,4 Dan-dan Wei,4 Yan-fang Mei,4 Dan Long,4 La-gen Wan,4 Yang Liu,4 Wei Zhang1
1Department of Respiratory and Critical Care Medicine, First Affiliated Hospital of Nanchang University, Nanchang University, Nanchang 330006, People’s Republic of China; 2The First Clinical Medical College of Nanchang University, Nanchang University, Nanchang 330006, People’s Republic of China; 3Department of Endocrinology and Metabolism, First Affiliated Hospital of Nanchang University, Nanchang University, Nanchang 330006, People’s Republic of China; 4Department of Clinical Microbiology, First Affiliated Hospital of Nanchang University, Nanchang University, Nanchang 330006, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yang Liu
Department of Clinical Microbiology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi 330006, People’s Republic of China
Tel +86-13576091584
Fax +86-0791-88692748
Email [email protected]
Wei Zhang
Department of Respiratory Medicine, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi 330006, People’s Republic of China
Tel +86-13707089183
Fax +86-0791-88692748
Email [email protected]
Introduction: Carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) is increasingly reported worldwide, but ceftazidime/avibactam (CAZ/AVI)-resistant hvKP isolates have rarely been observed. We attempted to characterize them in clinical CRKP isolates collected from a university hospital in China from March 2016 to March 2018.
Methods: All isolates were analyzed by antimicrobial susceptibility testing, molecular detection of antibiotic resistance determinants, multilocus sequence typing (MLST), SDS-PAGE, and pulsed-field gel electrophoresis (PFGE). The pLVPK-related genetic loci (rmpA2, terW, iutA, and silS) were screened in all CAZ/AVI-resistant CRKP isolates for the presence of virulence plasmids by PCR. Capsule typing, serum killing assay, Galleria mellonella lethality experiments, and mouse lethality assay were conducted to identify CAZ/AVI-resistant hvKP among isolates that carried all four virulence genes.
Results: A total of 232 CRKP isolates were collected. Overall, CAZ/AVI-resistance was found in 8.2% (19/232) CRKP isolates isolated from patients with no history of previous CAZ/AVI-based treatment. Among these, 63.2% (12/19) were metallo-β-lactamase-producing K. pneumoniae (MBL-KP), 52.6% (10/19) were Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae (KPC-KP), and 26.3% (5/19) produced both MBL and KPC. The presence of carbapenemase promoted a very high increase in CAZ/AVI minimum inhibitory concentration only when ompk35 and ompk36 were absent. Alarmingly, nine isolates had all four virulence genes for the presence of virulence plasmids. All nine isolates were considered to be CAZ/AVI-resistant hvKP according to the G. mellonella infection model and mouse lethality assay, with ST23 being the most common type (55.6%, 5/9).
Conclusion: The newly emerged hypervirulent CAZ/AVI-resistant KP strain might cause a serious threat to public health, suggesting an urgent need for enhanced clinical awareness and epidemiologic surveillance.
Keywords: ceftazidime/avibactam-resistant, hypervirulent, Klebsiella pneumoniae, ompk35
Introduction
The carbapenem-resistant Klebsiella pneumoniae (CR-KP) strain is associated with high morbidity and mortality—especially the carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) strain—and is therefore considered one of the most serious clinical threats to human health.1 In recent times, the continuous emergence of CR-hvKP has been increasingly reported predominantly from China.2 The complicated clinical practice caused by these CR-hvKP strains calls for the use of novel antibiotics to efficiently control these infections. The urgent need for new drugs with anti-CRKP activity has finally been addressed by the recent introduction of novel β-lactam/β-lactamase inhibitor combinations, among which ceftazidime-avibactam (CZA/AVI) was the first to be released for clinical use.3
Ceftazidime-avibactam is a new type of β-lactam/β-lactamase inhibitor combination that is effective against Enterobacteriaceae producing KPCs, OXA-48, carbapenemases, extended-spectrum β-lactamases, and AmpC β-lactamases, but it is not effective against those producing metallo-β-lactamases.4 A previous study has shown that CAZ/AVI provides a valuable alternative strategy against CR-hvKP, including the KPC-2-producing ST11 hvKP isolates, which are increasingly isolated in China.5 Although CAZ/AVI-based therapies have shown initial optimistic clinical efficacy in treating severe KPC-Kp infections, recent reports of development of drug resistance have been cause for concern.6,7 Previous studies showed that CAZ/AVI resistance was associated with different mutations in the KPC enzyme or overexpression of the blaKPC gene associated with non-functional porin.6,8 However, to date, clinical isolates of CR-hvKP resistant to CAZ/AVI have been rarely found. Therefore, this study aims to investigate the patterns of hypervirulence in CAZ/AVI-resistant CRKPs and further analyze the mechanisms of resistance.
Materials and Methods
Bacterial Isolates and Antimicrobial Susceptibility Tests
Between March 2016 and March 2018, 232 CR-KP isolates were isolated from patients hospitalized in a large tertiary hospital in Southern China. Antimicrobial susceptibility testing was performed using the VITEK II system (bioMérieux, Balmes-Les-Grottes, France) and confirmed by microdilution method. Results were interpreted following the Clinical and Laboratory Standards Institute (CLSI) guidelines.9 All isolates were characterized as resistant to imipenem or meropenem and were determined with phenotypic screening for carbapenemase production. The breakpoint of CAZ/AVI was based on the interpretative criteria according to K-B method of CLSI. Results of tigecycline and colistin were interpreted according to the breakpoint approved by the EUCAST.10 The reference strains—K. pneumoniae ATCC 700603 and Escherichia coli ATCC 25922—were used for quality control of antimicrobial susceptibility testing.
Capsular Serotyping, Resistance Genes, and pLVPK-Related Genes Detection
Polymerase chain reaction (PCR) was used to detect resistance genes related to carbapenemases (GES, KPC, NMC, SME, IMP, VIM, GIM, SPM, SIM, NDM, and OXA); β-lactamase (SHV, TEM, CTX-M, VEB, CMY, and DHA); and fenestra protein genes (ompK35 and ompK36) of the bacterial isolates. The eight capsular serotyping genes (K1, K2, K5, K20, K54, K57, K47, and K64) and pLVPK-related genetic loci (rmpA2, terW, iutA, silS) were also determined by PCR and sequencing as previously described.11–16
Outer Membrane Protein Isolation and SDS-PAGE
Outer membrane proteins (OMPs) were isolated according to the rapid procedure of Carlone et al and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described.17 The reference strains K. pneumoniae ATCC 700603 and K. pneumoniae NUTH-K2044 were also used as quality control.
G. mellonella Infection Model
All the clinical K. pneumoniae isolates were grown to the late exponential phase in MH broth; the isolates were then collected by centrifugation and suspended in saline. Ten larvae weighing between 250 and 350 mg (purchased from Tianjin Huiyude Biotech Company, Tianjin, China) were used for the assessment of the virulence level of each strain. The larvae were inoculated by injecting 1×106 CFU of clinical K. pneumoniae isolates per 10 microliters aliquot into the hemocoel via the rear left proleg using a 10-μL Hamilton animal syringe.15 Ten larvae were also injected with 1×106 CFU ATCC700603 and 1×106 CFU NTUH-K2044, respectively, as the controls. Larvae were incubated at 37°C and observed every 24 h for 5 days to monitor mortality. The average survival time of each group of infected larvae was recorded.
Serum Killing Assay
Serum killing assay was performed as described previously.18 Briefly, prior to the assay, serum separated from 10 healthy individuals’ blood was stored at −80°C. A 106 CFU of bacteria-containing inoculum prepared from the mid-log phase was reacted with 75% pooled human sera. The final mixture was incubated at 37°C, and we obtained viable counts at 0, 1, and 3 h, respectively. The response to serum killing in terms of viable counts was scored using six grades classified as serum sensitive (grade 1 or 2), intermediately sensitive (grade 3 or 4) or serum resistant (grade 5 or 6).
S1-PFGE and Southern Blot
After PCR screening of pLVPK-related genetic loci, S1 nuclease-pulsed-field gel electrophoresis (S1-PFGE) and southern blotting hybridization were performed to determine the location of virulence genes. Briefly, total DNA was embedded in agarose gel plugs. The plugs were digested with S1 nuclease (TaKaRa) at 37°C for 30 min and then separated by electrophoresis. Labeling of the probe rmpA2 and hybridization were performed with the DIG-High Prime DNA Labeling and Detection Starter Kit II, according to the manufacturer’s instructions (Roche, Basel, Switzerland).19
Mouse Lethality Assay
Determination of the virulence of KP in mouse lethality tests and the medium lethal dose (LD50, expressed as colony-forming units) was performed as previously described.20 In brief, a graded dose of 101 to 106 CFU of each strain in 10-fold serial dilutions in 0.1 mL of normal saline was injected intraperitoneally into mice (4 mice for each dose of inoculum). All inoculated mice were recorded daily for survival. Interpretation of virulence was referred to reference.21
Multilocus Sequence Typing (MLST) and PFGE
Seven conserved housekeeping genes—gapA, infB, mdh, pgi, phoE, rpoB, and tonB—were used to perform MLST (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). Clonal relatedness among CAZ/AVI-resistant K. pneumoniae isolates was established using XbaI - PFGE (TaKaRa). DNA fragments were separated with a CHEF DR III apparatus (Bio-Rad, Richmond, CA, USA). The molecular marker was Salmonella serotype Braenderup strain H9812. The isolates sharing >80% similarity were defined as the same PFGE cluster.22
Ethics Statement
The study has been evaluated by the Ethics Committee of the First Affiliated Hospital of Nanchang University (ethical number: 2019130). Patients involved in the study were anonymized, and the need for informed consent was waived because of the retrospective nature of the study.
Results
Prevalence of Carbapenemase Genes and β-Lactamase Genes Among CAZ/AVI-Resistant K. pneumoniae Clinical Isolates
A total of 232 carbapenem-resistant K. pneumoniae clinical isolates were collected from March 2016 to March 2018 in our hospital. Among these, 19 CAZ/AVI-resistant K. pneumoniae clinical isolates were selected from clinical specimens including 10 sputum, 4 blood, 3 urine, and 2 pus. As shown in Supplementary Table 1, almost all the CAZ/AVI-resistant K. pneumoniae clinical isolates were resistant to 20 antibiotics commonly used in clinical treatment, except for some isolates that were sensitive to tigecycline, polymyxin, and amikacin. Hence, these CAZ/AVI-resistant K. pneumoniae clinical isolates, particularly CAZ/AVI-resistant hvKP isolates, understandably pose massive challenges for clinical treatment. As shown in Table 1, almost all of the CAZ/AVI-resistant K. pneumoniae clinical isolates were found to carry at least one carbapenemase gene or extended-spectrum β-lactamase gene. Interestingly, among these CAZ/AVI-resistant K. pneumoniae clinical isolates, NDM-1/NDM-5 were more likely clustered in hvKP in our hospital.
 |
Table 1 Virulence Phenotype of CAZ/AVI-Resistant K. pneumoniae Isolates and Drug-Resistant Genes |
Virulence Assessment of CAZ/AVI-Resistant K. pneumoniae Clinical Isolates
In this scenario, capsular serotyping showed there were 10 K46 isolates, five K1 isolates, two K2 isolates, one K14 isolate, and one K15 isolate among CAZ/AVI-resistant K. pneumoniae clinical isolates. PCR analysis of pLVPK-related genetic loci revealed there were nine CAZ/AVI-resistant K. pneumoniae isolates that may carry the pLVPK-like virulence plasmid. These results were verified by PFGE and Southern blotting. As shown in Supplemental Figure 1, nine (47.4%) CAZ/AVI-resistant hvKP clinical isolates were identified among 19 CAZ/AVI-resistant K. pneumoniae clinical isolates. Moreover, the serum killing assay, G. mellonella infection model, and mouse lethality assay also proved that the nine CAZ/AVI-resistant hvKP clinical isolates were hypervirulent.
SDS-PAGE of Outer Membrane Proteins
The outer membrane proteins of 19 CAZ/AVI-resistant K. pneumoniae clinical isolates were detected by SDS-PAGE. Two ST11 isolates lacked outer membrane proteins, as shown in Figure 1. PCR analysis of the outer membrane protein genes also revealed absence of the OmpK35 and OmpK36 genes. Therefore, unlike the other seven CAZ/AVI-resistant hvKP isolates carrying the NDM gene, these two ST11 hvKP isolates were resistant to CAZ/AVI because of loss of the outer membrane proteins.
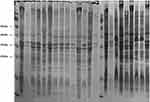 |
Figure 1 SDS-PAGE of outer membrane proteins of 19 clinical CAZ/AVI-resistant K. pneumoniae isolates. Note: the dotted rectangle reveals the absence of Ompk35 and Ompk36. |
Molecular Characteristics
Among the 19 CAZ/AVI-resistant K. pneumoniae isolates, five STs were identified, including ST11 (10 K64 isolates), ST23 (five K1 isolates), ST86 (two K2 isolates), ST345 (one K14 isolate), and ST4065 (one K25 isolate). The PFGE patterns were assigned to four clusters based on >80% pattern similarity, comprising seven ST11 isolates, five ST23 isolates, three ST11 isolates, and two ST86 isolates (Figure 2). The remaining two isolates (ST14 and ST25) had different pulsotypes. The PFGE and MLST results suggested that CAZ/AVI-resistant hvKP isolates mainly clustered in K1 ST23 isolates and K2 ST86 isolates.
 |
Figure 2 Gel electrophoresis (PFGE) patterns, capsular genotypes, and STs of the 19 clinical CAZ/AVI-resistant K. pneumoniae isolates. |
Discussion
The use of CAZ/AVI against carbapenem-resistant K. pneumoniae strains has been shown to be very effective to provide a higher clinical cure rate and survival rate in China, the United States, and other European countries.23–26 However, increased reports of CAZ/AVI resistance in carbapenem-resistant K. pneumoniae, regardless of prior exposure, is a matter of concern.27–29 A national research about CAZ/AVI resistance in China revealed that MBL production, blaKPC-2 point mutation and high KPC expression played an important role in CAZ/AVI resistance.28 Nevertheless, reports of CR-hvKP with CAZ/AVI resistance have been rare.
Here, we described the emergence of 19 CAZ/AVI-resistant carbapenem-resistant K. pneumoniae isolates including nine CR-hvKP isolates isolated from patients with no history of previous antimicrobial exposure to CAZ/AVI treatment. Our results showed that CAZ/AVI resistance was mainly associated with the presence of metallo-β-lactamases or deficiency of OmpK35/36 porins. Although previous studies have demonstrated that the mechanisms responsible for CAZ/AVI resistance included specific mutations within the blaKPC gene,6,30 PCR analysis of the KPC gene in this study showed no mutations in the KPC-3 gene. As described in a previous study,31 CAZ/AVI resistance and KPC-3 amino acid substitutions only occurred at low frequencies (~10−9) in K. pneumoniae during in vitro selection. Moreover, due to the limited use of CAZ and AVI in our hospital, the probability of genetic mutations was further reduced.
SDS-PAGE of the outer membrane protein in this study revealed that only two ST11 isolates that carried the pLVPK-like virulence plasmids lacked the outer membrane proteins. Therefore, the loss of porin OmpK35/36 was not a primary and direct cause of resistance to CAZ/AVI in this study. Per previous studies, the OmpK35 and OmpK36 porins were not thought to be the initial pathway for AVI into K. pneumoniae,32 rather the combination of this with increased KPC expression or production of DHA-1 β-lactamases could likely have led to the CAZ/AVI resistance phenotype.33,34
Importantly, nine (47.4%) CAZ/AVI-resistant hvKP clinical isolates carrying pLVPK-like virulence plasmids were identified in this study. The serum killing assay and G. mellonella infection model and mouse lethality assay also proved that the nine CAZ/AVI-resistant hvKP clinical isolates were more virulent than the remaining CAZ/AVI-resistant clinical isolates. Although Yu et al5 found that CAZ/AVI remained highly active against KPC-2-producing ST11 hvKp isolates, two CAZ/AVI-resistant ST11 CR-hvKP clinical isolates had first emerged in our hospital because of porin OmpK35/36 loss and DHA-1 β-lactamases production.
The PFGE and MLST results of the 19 CAZ/AVI-resistant K. pneumoniae isolates revealed that the CAZ/AVI-resistant hvKP in this study were the ST23 (K1)/ST86 (K2) isolates carrying the NDM gene and the ST11 KPC-hvKP isolates that lacked OmpK35/36 porins and DHA-1 β-lactamases production. Both these types of CAZ/AVI-resistant hvKP isolates need further monitoring.
Our study has some limitations, including its retrospective nature and a relatively small study population. Therefore, there may be selection bias, which limits the applicability and generalizability of our results to other areas. Although we did not discuss efflux pump mechanisms of CAZ/AVI resistance, to our best knowledge, this is the first description of CAZ/AVI resistance in carbapenem-resistant hypervirulent Klebsiella pneumoniae.
Conclusion
The major mechanisms of CAZ/AVI resistance among carbapenem-resistant Klebsiella pneumoniae in our hospital were metallo-β-lactamases production and loss of porin OmpK35/36 along with DHA-1 β-lactamases production. Effective infection control measures are urgently needed to prevent the emerging hypervirulent CAZ/AVI-resistant hvKP strains from becoming an epidemic in the future.
Abbreviations
CR-hvKP, carbapenem-resistant hypervirulent Klebsiella pneumoniae; CAZ/AVI, ceftazidime/avibactam; MLST, multilocus sequence typing; PFGE, pulsed-field gel electrophoresis; MBL-KP, metallo-β-lactamase-producing K. pneumoniae; KPC-KP, Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae; G. mellonella, Galleria mellonella; CR-KP, carbapenem-resistant Klebsiella pneumoniae; S1-PFGE, S1 nuclease-pulsed-field gel electrophoresis; CLSI, the Clinical and Laboratory Standards Institute; PCR, polymerase chain reaction; OMPs: outer membrane proteins; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; LD50, the medium lethal dose.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–81. doi:10.1073/pnas.1501049112
2. Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. doi:10.1093/jac/dkz446
3. Shirley M. Ceftazidime-Avibactam: A Review in the Treatment of Serious Gram-Negative Bacterial Infections. Drugs. 2018;78(6):675–692. doi:10.1007/s40265-018-0902-x
4. van Duin D, Bonomo RA. Ceftazidime/Avibactam and Ceftolozane/Tazobactam: second-generation β-Lactam/β-Lactamase Inhibitor Combinations. Clin Infect Dis. 2016;63(2):234–241. doi:10.1093/cid/ciw243
5. Yu F, Lv J, Niu S, et al. In vitro activity of ceftazidime-avibactam against carbapenem-resistant and hypervirulent Klebsiella pneumoniae isolates. Antimicrob Agents Chemother. 2018;62:8. doi:10.1128/AAC.01031-18
6. Shields RK, Chen L, Cheng S, et al. Emergence of Ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant klebsiella pneumoniae infections. Antimicrob Agents Chemother. 2017;61:3. doi:10.1128/AAC.02097-16
7. Gaibani P, Campoli C, Lewis RE, et al. In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J Antimicrob Chemother. 2018;73(6):1525–1529. doi:10.1093/jac/dky082
8. Nelson K, Hemarajata P, Sun D, et al. Resistance to Ceftazidime-Avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother. 2017;61:10. doi:10.1128/AAC.00989-17
9. Humphries RM, Ambler J, Mitchell SL, et al. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol. 2018;56(4):4. doi:10.1128/JCM.01934-17
10. EUCAST. 2018. Breakpoint tables for interpretation of MICs and zone diameters. Version 8.0, January 2018. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.pdf.
11. Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–58, table of contents. doi:10.1128/CMR.00001-07
12. Turton JF, Perry C, Elgohari S, Hampton CV. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59(Pt 5):541–547. doi:10.1099/jmm.0.015198-0
13. Brisse S, Passet V, Haugaard AB, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51(12):4073–4078. doi:10.1128/JCM.01924-13
14. Wassef M, Abdelhaleim M, AbdulRahman E, Ghaith D. The role of OmpK35, OmpK36 Porins, and Production of β-lactamases on imipenem susceptibility in Klebsiella pneumoniae clinical isolates, Cairo, Egypt. Microb Drug Resist. 2015;21(6):577–580. doi:10.1089/mdr.2014.0226
15. Liao W, Liu Y, Chen C, et al. Distribution of CRISPR-Cas systems in clinical carbapenem-resistant Klebsiella pneumoniae strains in a Chinese Tertiary Hospital and its potential relationship with virulence. Microb Drug Resist. 2019. doi:10.1089/mdr.2019.0276
16. Makowska N, Philips A, Dabert M, et al. Metagenomic analysis of β-lactamase and carbapenemase genes in the wastewater resistome. Water Res. 2020;170:115277. doi:10.1016/j.watres.2019.115277
17. Hao M, Ye M, Shen Z, et al. Porin Deficiency in Carbapenem-Resistant Enterobacter aerogenes Strains. Microb Drug Resist. 2018;24(9):1277–1283. doi:10.1089/mdr.2017.0379
18. Liu Y, Long D, Xiang TX, et al. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J Antimicrob Chemother. 2019;74(5):1233–1240. doi:10.1093/jac/dkz023
19. Xu M, Fu Y, Fang Y, et al. High prevalence of KPC-2-producing hypervirulent Klebsiella pneumoniae causing meningitis in Eastern China. Infect Drug Resist. 2019;12:641–653. doi:10.2147/IDR.S191892
20. Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62(1):1–6. doi:10.1016/j.diagmicrobio.2008.04.007
21. Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12(11):881–887. doi:10.1016/S1473-3099(12)70205-0
22. Li J, Huang ZY, Yu T, et al. Isolation and characterization of a sequence type 25 carbapenem-resistant hypervirulent Klebsiella pneumoniae from the mid-south region of China. BMC Microbiol. 2019;19(1):219. doi:10.1186/s12866-019-1593-5
23. Farrell DJ, Sader HS, Flamm RK, Jones RN. Ceftolozane/tazobactam activity tested against Gram-negative bacterial isolates from hospitalised patients with pneumonia in US and European medical centres (2012). Int J Antimicrob Agents. 2014;43(6):533–539. doi:10.1016/j.ijantimicag.2014.01.032
24. Testa R, Cantón R, Giani T, et al. In vitro activity of ceftazidime, ceftaroline and aztreonam alone and in combination with avibactam against European Gram-negative and Gram-positive clinical isolates. Int J Antimicrob Agents. 2015;45(6):641–646. doi:10.1016/j.ijantimicag.2014.12.033
25. Shields RK, Nguyen MH, Chen L, et al. Ceftazidime-Avibactam is superior to other treatment regimens against carbapenem-resistant klebsiella pneumoniae bacteremia. Antimicrob Agents Chemother. 2017;61:8. doi:10.1128/AAC.00883-17
26. Yin D, Wu S, Yang Y, et al. Results from the China Antimicrobial Surveillance Network (CHINET) in 2017 of the in vitro activities of ceftazidime-avibactam and ceftolozane-tazobactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;63:4. doi:10.1128/AAC.02431-18
27. Giddins MJ, Macesic N, Annavajhala MK, et al. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-Harboring Klebsiella pneumoniae sequence Type 307 isolates. Antimicrob Agents Chemother. 2018;62:3. doi:10.1128/AAC.02101-17
28. Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):
29. Wei J, Zou C, Wang D, Huang A, Niu S. Genetic diversity and in vitro activity of ceftazidime/avibactam and aztreonam/avibactam against imipenem-resistant Enterobacteriaceae isolates in Southwest China: a single-centre study. J Glob Antimicrob Resist. 2020;22:448–451. doi:10.1016/j.jgar.2020.04.023
30. Venditti C, Nisii C, D’Arezzo S, et al. Molecular and phenotypical characterization of two cases of antibiotic-driven ceftazidime-avibactam resistance in blaKPC-3-harboring Klebsiella pneumoniae. Infect Drug Resist. 2019;12:1935–1940. doi:10.2147/IDR.S207993
31. Livermore DM, Warner M, Jamrozy D, et al. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother. 2015;59(9):5324–5330. doi:10.1128/AAC.00678-15
32. Pagès JM, Peslier S, Keating TA, Lavigne JP, Nichols WW. Role of the outer membrane and porins in susceptibility of β-lactamase-producing enterobacteriaceae to ceftazidime-avibactam. Antimicrob Agents Chemother. 2015;60(3):1349–1359. doi:10.1128/AAC.01585-15
33. Shi W, Li K, Ji Y, et al. Carbapenem and cefoxitin resistance of Klebsiella pneumoniae strains associated with porin OmpK36 loss and DHA-1 β-lactamase production. Braz J Microbiol. 2013;44(2):435–442. doi:10.1590/S1517-83822013000200015
34. Zhang Y, Jiang X, Wang Y, et al. Contribution of β-lactamases and porin proteins OmpK35 and OmpK36 to carbapenem resistance in clinical isolates of KPC-2-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2014;58(2):1214–1217. doi:10.1128/AAC.02045-12
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
