Back to Journals » Infection and Drug Resistance » Volume 15
Emergence of Ceftazidime–Avibactam Resistance and Decreased Virulence in Carbapenem-Resistant ST11 Klebsiella pneumoniae During Antibiotics Treatment
Authors Xu M, Qian C, Jia H, Feng L, Shi S, Zhang Y, Wang L, Cao J, Zhou T , Zhou C
Received 23 September 2022
Accepted for publication 23 November 2022
Published 28 November 2022 Volume 2022:15 Pages 6881—6891
DOI https://doi.org/10.2147/IDR.S387187
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Mengxin Xu,1 Changrui Qian,2 Huaiyu Jia,1 Luozhu Feng,3 Shiyi Shi,1 Ying Zhang,3 Lingbo Wang,1 Jianming Cao,3 Tieli Zhou,1 Cui Zhou1
1Department of Clinical Laboratory, the First Affiliated Hospital of Wenzhou Medical University, Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province, Wenzhou, People’s Republic of China; 2School of Basic Medical Sciences, Wenzhou Medical University, Wenzhou, People’s Republic of China; 3Department of Medical Laboratory Science, School of Laboratory Medicine and Life Science, Wenzhou Medical University, Wenzhou, People’s Republic of China
Correspondence: Tieli Zhou; Cui Zhou, Department of Clinical Laboratory, the First Affiliated Hospital of Wenzhou Medical University; Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province, Wenzhou, 325035, People’s Republic of China, Tel +86 0577 8668 9885, Email [email protected]; [email protected]
Introduction: Carbapenem-resistant Klebsiella pneumoniae (CRKP) poses a serious threat to human public health. Ceftazidime–avibactam (CZA) is currently one of the few effective antibiotics for carbapenem-resistant Enterobacteriaceae (CRE).
Methods and Results: Here, we analyzed two longitudinal Klebsiella pneumoniae clinical isolates (FK8578, FK8695) that were isolated from an ICU patient during antimicrobial treatment. Broth microdilution method, whole-genome sequencing (WGS) and comparative genomic analysis were used to elucidate the dynamics and mechanisms of antibiotic resistance. String test, quantification of capsule, biofilm inhibition test and Galleria mellonella (G. mellonella) infection model were used to explore the changes in virulence of the two clinical isolates. During antibiotic treatment, CRKP FK8578 underwent a series of drug resistance and virulence changes, including CZA resistance, carbapenem susceptibility and virulence attenuation. The results of WGS showed that mutation of blaKPC-2 to blaKPC-33 was responsible for the change of drug resistance phenotype between FK8578 and FK8695. pLVPK-like virulence plasmid without siderophore synthesis operon was identified in the two strains. On the other hand, the loss of hypermucoviscosity phenotype in the FK8695 strain may be related to a single nucleotide deletion of the rmpA gene, which would further lead to a decrease in virulence. Virulence results showed that compared with FK8578, FK8695 was negative in the string test, with decreased capsular production, smaller amounts of biofilm formation and higher survival rate of G. mellonella.
Conclusion: This is the first report of CZA resistance and decreased virulence in ST11 CRKP strains during antimicrobial treatment. It is urgent to monitor CZA resistance and timely adjust anti-infective treatment strategies.
Keywords: ceftazidime–avibactam, carbapenem-resistantKlebsiella pneumoniae, whole-genome sequencing, hypermucoviscosity, pLVPK-like plasmid, virulence attenuation
Introduction
Owing to the extensive use of carbapenems in clinical practice, the prevalence of carbapenem-resistant Klebsiella pneumoniae (CRKP) increased globally, which poses a serious challenge to clinical treatment.1 CZA is a novel β-lactam and β-lactamase inhibitor combination with the ability to inhibit the activity of AmpC, extended-spectrum β-lactamases (ESBLs), class A carbapenemases such as KPC and class D carbapenemases such as OXA-48 but not class B carbapenemases such as NDM, IMP and VIM2,3 and has been considered as a last-resort treatment for severe infections caused by carbapenem-resistant Enterobacterales (CRE). Since the use of CZA was approved by the United States in 2015, the European Union in 2016, and China in 2019, the occurrence of CZA resistance has been increasingly reported worldwide.4,5
In addition to CRKP, hypervirulent K. pneumoniae (hvKp) is another threat that causes severe community-acquired infections. The hypervirulence of K. pneumoniae was contributed to a virulence plasmid pLVPK which harbors capsular polysaccharides regulator genes rmpA/rmpA2 and some siderophore determinants.6 Hypermucoviscosity (Hmv) phenotype is usually associated with hvKp, which was caused by the production of capsular polysaccharide (CPS) and the presence of virulence genes such as rmpACD/rmpA2 and magA.7–9 Recently, the convergence of virulence and carbapenem resistance in K. pneumoniae has been reported, especially in China.10 Carbapenem-resistant hvKp (CR-hvKp) strains mainly belong to the sequence type 11 (ST11) and capsular serotype K47/K64, which originated from the ST11 CRKP obtaining a pLVPK-like virulence plasmid and majority of HvKP belonged to capsular serotype K1 such as ST23, and capsular serotype K2 including ST86, ST65, ST25, and ST375.11 The superbug has the potential to trigger the next clinical crisis and cause severe, untreatable infections in previously healthy people.
Ceftazidime–avibactam (CZA) has been used as a last-resort option for the treatment of serious infections caused by carbapenem-resistant Enterobacterales producing either KPC or oxacillinase 48 (OXA-48)-like carbapenemases, being ineffective against producers of metallo-β-lactamases.12 However, clinical isolates of ST11 CRKP resistant to CZA have been rarely described.13 In this study, we elucidated the antimicrobial resistance and virulence changes of ST11-CRKP using whole-genome sequencing (WGS) and bioinformatics analysis. The ST11-CRKP strain was longitudinally isolated from ascites samples obtained from patients during 14 days course of CZA-containing antibiotic therapy. In this process, ST11-CRKP underwent a series of phenotypic variations, including resistance to CZA, susceptibility to carbapenems, decreased production of capsular polysaccharides (CPS), decreased number of biofilm formation, and improved survival rate of G. mellonella. In this study, we report for the first time the emergence of CZA resistance in ST11 CRKP during CZA treatment. Importantly, we observed attenuation of virulence accompanied by acquisition of antibiotic resistance.
Materials and Methods
Bacterial Isolates and Identification
In 2020, a patient was hospitalized in the ICU for septic shock, head trauma, hydrocephalus (after surgery) and other diseases. He underwent a lumbar puncture on the day of admission and was temporarily given meropenem to anti-infective. On the second day, the patient had difficulty breathing suddenly, and emergency endotracheal intubation and ventilator-assisted ventilation were performed, while gram-positive bacteria were covered with linezolid. On day 5, the patient’s blood culture revealed K. pneumoniae infection. CZA was used instead of meropenem and linezolid for anti-infection. On day 10, the patient underwent peritoneal puncture. The first strain of CRKP (FK8578, blaKPC-2 positive) was isolated from ascitic fluid. The patient’s condition was stable and transferred to the hepatobiliary surgery ward. After the patient was treated with CZA for 14 days, FK8695 was isolated from the patient’s ascitic fluid, during which time K. pneumoniae was isolated from the patient repeatedly. FK8578 and FK8695 were isolated from the patients before and after the drug sensitivity of CZA and carbapenems was changed. The CZA-resistant strain FK8695 and the progenitor strain FK8578 were identified at the species level using a VITEK®MS automated system (BioMérieux, Hazelwood, MO, USA).
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of clinical routine antimicrobial agents were performed by broth microdilution method.14 Clinical routine antimicrobial agents such as ceftazidime–avibactam, ertapenem, meropenem, imipenem, ceftazidime, cefepime, ceftriaxone, levofloxacin, amikacin, gentamicin, colistin, tigecycline. Escherichia coli ATCC 25922 were employed as reference strains. The results were interpreted according to 2020 CLSI breakpoints,15 except for tigecycline and colistin, which were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines.16 All experiments are repeated three times.
String Test
The FK8578 and FK8695 were inoculated on 5% sheep blood plates and cultured overnight at 37℃. The colony was pulled up with an inoculation loop to form a line. Hmv was defined as the formation of viscous strings >5 mm in length (positive string test) when the colony was stretched on the plate with a ring.17
Extraction and Quantification of Capsule
The extraction and quantification of uronic acid were performed as previously described, with some modifications.18,19 Briefly, the culture was grown to an OD600 of 0.2 in fresh media. After 6 h, the adjusted OD600 of the culture was 1.0 mL−1. Five hundred microliters of the bacterial suspension was mixed with 100 µL 1% zwittergent 3–14 detergent in citric acid, then incubated for 20 min at 50℃. After centrifugation (14,000 rpm for 2 min). Next, 300 µL of supernatant were added to 1.2 mL of absolute ethanol and then were centrifuged (14,000rpm, 5min). After resuspension of the pellet in 200 µL of distilled water, to which 1.2 mL sodium tetraborate in sulfuric acid was mixed, the solution was boiled for 5 min and stored on ice for 10 min. 3-Phenylphenol in 0.5% NaOH was added. After 5 minutes, the absorbance was measured at 520 nm. The glucuronic acid content was analyzed from a standard curve of glucuronic acid and expressed as µg/108CFU.
Biofilm Formation Assays
The biofilm biomass was analyzed using crystal violet staining, and some modifications were made.20 Briefly, the culture was grown to 0.5 McFarland, diluted with sterile saline in LB broth, and placed in 96-well microtiter plates. After incubation at 37℃ for 24 h, the floating and non-adherent cells were removed by washing them twice with phosphate-buffered saline (PBS). Then, let it dry at room temperature. Next, 200 µL of 1% crystal violet (CV) solution (Beijing Solarbio Biotechnology Co., Ltd., China) was added for each well, incubated for 15 minutes, removed CV after staining, and washed 2–3 times with 1× PBS. Two hundred microliters of 95% v/v ethanol (95% absolute ethanol and 5% glacial acetic acid) was dissolved and stained the biofilm at 37℃ for 10 min. The absorbance was measured in a microplate reader at 595 nm (Multiskan FC). The experiments were repeated three times.
Galleria mellonella Infection Model
The changes in virulence were evaluated by the G. mellonella infection in vivo model with minor modification.21 Milky-white G. mellonella larvae with a body weight of 250–300 mg were selected for the experiment. We used 10 G. mellonella in each group. We used 70% alcohol to surface sterilize Galleria mellonella larvae before the bacteria were injected. Firstly, bacteria were inoculated on blood plates and cultured overnight for 18 to 24h. A single colony was selected from the blood plate and adjusted to 0.5 McFarland with sterile NS. Then, the bacterial suspension was further diluted to 1.5×107 CFU/mL. Next, G. mellonella was injected with 10 μL of bacterial suspension into the last left proleg using a microsyringe. The negative control group received only sterile saline. The experimental group was injected with the corresponding concentration of bacterial suspension. The larvae were placed at 37℃ and their survival rate was recorded for 7 days. Larval death was mainly determined by changes in skin color and lack of response to stimuli. Kaplan–Meier method and Log rank test were used to analyze the larval survival rate. All experiments were repeated three times.
Whole-Genome Sequencing (WGS) and Bioinformatics Analysis
The genomic DNA of FK8578 and FK8695 was extracted using an AxyPrep bacterial genomic DNA miniprep kit (Axygen Scientific, Union City, CA, USA), and subsequently sequenced using a standard run of Illumina HiSeq 2500. FK8695 was further sequenced by Pacific Bioscience (PacBio) systems (Shanghai Personal Biotechnology Co., Ltd. Shanghai, China). Complete genomes were assembled by Canu v2.022 and corrected by Pilon v1.2423 with default parameters. Genome sequences were annotated using Prokka v1.14.524 combined with ISfinder23 (https://www-is.biotoul.fr/), ResFinder25 (https://cge.cbs.dtu.dk/services/ResFinder/) and Kleborate v2.0.426 with default parameters. Variants calling between FK8578 and FK8695 were performed using Snippy v4.6 against the complete FK8578 genome using default parameters. Plasmid circular map was generated using CGview v2.02.27 Gene organization diagrams were generated using Python script and edited with Inkscape v1.2 (https://inkscape.org/en/).
Conjugation Experiments
The conjugation experiments were used with rifampicin-resistant E. coli EC600 as a recipient strain to explore the transferability of CZA resistance. Briefly, the donor and recipient strain were cultured in Luria broth medium and incubated overnight at 37℃ with shaking. Then, a dilution of 1/50 of each donor and recipient strain was made on fresh Luria broth medium (10 mL) and further incubated for 4h at 37℃ with shaking. A volume of 1 mL of donor cells and 4 mL of recipient cells was mixed together and incubated for 2h at 37℃ without shaking. Transconjugants were screened on Luria–Bertani agar plates containing ceftazidime–avibactam (16 mg/L) and rifampicin (600 mg/L). The plates were incubated overnight at 37℃ for 16–18 h. PCR and drug susceptibility testing were performed to verify the presence of blaKPC. Primers: (KPC-F: 5′- AAGATCTACAACCACAGCATTC- 3′; KPC-R: 5′- CAGACTCCTAGCCTAAATGTGA- 3′).
Access to Data
The genome data of FK8578 and FK8695 have been submitted to NCBI under BioProject accession number PRJNA774952.
Statistical Analysis
The results were expressed as mean ± standard deviation. The significance was determined using Student t-test and Log rank test. For all analyses: *P < 0.05, **P < 0.01, and ***P < 0.001. Statistical analysis was performed with Prism 8.
Results
Isolate Characterizations
The results of antimicrobial susceptible testing showed that K. pneumoniae FK8578 and FK8695 were resistant to ceftazidime, cefepime, ceftriaxone, and levofloxacin; however, it was sensitive to amikacin, gentamicin, colistin, and tigecycline (Table 1). In addition, K. pneumoniae FK8695 showed increased CZA-resistance but reduced carbapenem resistance compared with K. pneumoniae FK8578. Clinical and microbiological details, timelines, and antibiotic treatments used are shown in Figure 1.
 |
Table 1 MIC Values of Commonly Used Clinical Antibiotics Against FK8578 and FK8695 |
 |
Figure 1 Time courses of infection and treatment among a patient in whom CZA-resistant K. pneumoniae emerged. |
Decreased Hmv Phenotype and CPS Formation in CZA-Resistant Strain
The Hmv phenotype was observed in the two K. pneumoniae strains by string test. The CZA-susceptible strain FK8578 showed a positive string test, whereas the clinical CZA-resistant strain FK8695 showed a negative string test (Figure 2A). Consistent with these results, CPS production by the CZA-resistant FK8695 strain decreased compared with that of the CZA-susceptible FK8578 strain (Figure 2B).
Reduced Biofilm Formation and Improved G. mellonella Survival Rate
To assess the difference in virulence more closely, Figure 3A depicts the biofilm formation assay results of the two K. pneumoniae strains. CZA-susceptible strain FK8578 produced relatively robust biofilms (OD595, 3.63). However, CZA-resistant strain FK8695 had less biofilm formation. Consistent with these results, the virulence levels of FK8578 and FK8695 were further examined in the G. mellonella infection model. After G. mellonella infected with FK8578 (1×107 CFU/mL) for 24 h, the survival rate of larvae had reached to 30%. However, 70% survival was recorded for FK8695 (Figure 3B), suggesting that FK8578 was phenotypically highly virulent. These results indicated that the CZA-resistant strain FK8695 loss of Hmv and exhibited lower virulence compared with FK8578.
Whole-Genome Sequencing and Analysis of Antibiotic Resistome
In order to explore the difference of drug resistance and virulence of FK8578 and FK8695. WGS was performed on FK8578 and FK8695 to genetically characterize and explain their resistant phenotype. The complete genome of FK8695 consists of a 5.4 Mbp chromosome and four circular plasmids. The genomic features of FK8695 are presented in Table 2. FK8578 and FK8695 belonged to sequence type ST11 with capsular type KL64 based on in silico analysis. The sequence reads of FK8578 were mapped to the complete genome of the FK8695, and four mutations were identified between them (Table S1). Moreover, by mapping the assembled contigs of FK8578 to the complete plasmid sequences of FK8695, we found that the matching coverage of the two reached 100% (Figure S1). These results indicated that FK8695 was evolved from FK8578 and the plasmids sequences of the two strains were nearly identical.
 |
Table 2 General Features of FK8695 |
Twelve antimicrobial resistance genes (ARGs) were identified on FK8578 conferring resistance to aminoglycoside (aadA2b), β-lactam (blaLAP-2, blaKPC-2, blaCTX-M-65, blaSHV-11, blaSHV-12), fosfomycin (fosA), phenicol (catA2), fluoroquinolone (qnrS1), sulphonamide (sul2), tetracycline (tetA), and trimethoprim (dfrA14). The resistance genotype between FK8578 and FK8695 was nearly identical except that a blaKPC-33 rather than blaKPC-2 was identified in FK8695. KPC-33 was a KPC-2 variant with D179Y mutation in the omega loop, which can elevate CZA resistance, restore susceptibility to imipenem and low-level resistance to meropenem. In addition, blaKPC-33 gene was successfully transferred into E. coli EC600 and confirmed by PCR. Antimicrobial susceptibility testing results showed that the transconjugant was resistant to CZA, ertapenem, quinolone, and some cephalosporins (Table 3). Moreover, we examined the mutation of porin genes (ompK35, ompK36, and ompK37). We found that ompK36 was absent in two strains, while ompK35 and ompK37 of two strains were truncated by nonsense mutation (data not shown). These results indicated that the mutation of KPC-2 was responsible for the resistance of FK8695 to CZA.
 |
Table 3 Minimal Inhibitory Concentrations (MICs) of FK8578, FK8695 and the blaKPC-33-Positive E. coli Transconjugant of FK8695 |
Genetic Context of blaKPC-33 Gene
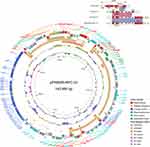
The blaKPC-33-harboring plasmid of FK8695, designated pFK8695-KPC-33, was 143,980 bp in length with an average G+C content of 53.76% (Figure 4). The plasmid contained two replicons, IncFII (pHN7A8) and IncR, and consisted of replication, maintenance, conjugation transfer and variable regions. Two ARGs blaCTX-M-65 and blaSHV-12 were identified in addition to blaKPC-33. Comparative genomic analysis of pFK8695-KPC-33 with other three blaKPC-33-harboring plasmid retrieved from GenBank revealed that they were belong to different incompatibility group (Figure 4). Among these blaKPC-33-harboring plasmid, pFK8695-KPC-33 shared the highest similarity (99.99% identity and 60% coverage) with the plasmid CP050351. The genetic context of blaKPC-33 on pFK8695-KPC-33 was a typical structure IS26-ISKpn27-blaKPC-33-TnAs1, which was identical to that of plasmid MT550691 (Figure 4) but was different with plasmid CP050351 (IS26-ISKpn27-blaKPC-33-IS26) and plasmid CP025144 (Tn4401).
Comparative Analysis of Virulence Determinants
As the ST11 CRKP strain, we were surprised to find that FK8578 and FK8695 also carried a virulence plasmid. The virulence plasmid of FK8695, designated pFK8695-rmpA, was 150,052 bp in length with an average G+C content of 50.04%. The plasmid belonged to IncHI1B /repB group and shared the highest similarity (99.99% identity and 99.97% coverage) with plasmid pVir-CR-hvKP-C789 (CP034416) from a KPC-2-producing K64-ST11 CR-hvKP strains in GenBank. In general, these virulence plasmids carried putative virulence factors such as aerobactin synthesis operon (iucABCD), the outer membrane ferric aerobactin receptor (iutA), a gene cluster of salmochelin production (iroBCDN), and regulator of mucoid phenotype A (rmpACD and/or rmpA2).6 Notably, the gene clusters iucABCD-iutA and iroBCDN were absent in pFK8695-rmpA (Figure 5). These two clusters were located on typical virulence plasmid pLVPK (AY378100) and encoded for siderophore aerobactin and salmochelin, respectively. Loss of iucABCD-iutA gene clusters resulted in a truncated iroN gene in pFK8695-rmpA and pVir-CR-hvKP-C789.
Virulence factors were detected of FK8578 and FK8695 to explain the hypermucoviscosity and virulence phenotype, but no genotypic differences were observed. We found that two strains have the same number and type of virulence determinants both in chromosome and plasmids. However, a frameshift mutation that deleted one nucleotide at the poly (G) region of rmpA gene was observed in FK8695 (Table S1). Since rmpA2 was not identified both in FK8578 and FK8695, inactivation of the rmpA gene in FK8695 might be responsible for the altered mucoviscosity; this further leads to a reduction in virulence.
Discussion
The emergence of CZA resistance in CRKP has resulted in limited effective treatment strategies and poses a great challenge to public health security. What is worse, the convergence of carbapenem-resistant and hypervirulent in K. pneumoniae cause severe and untreatable invasive infections. In this study, we report the emergence of resistance to CZA and the loss of Hmv in ST11 CR-hvKp during antimicrobial therapy.
It has been reported that CZA resistance is associated with specific mutations of blaKPC, blaCTX-M, and outer membrane protein (omp) genes, increased copy number of blaKPC variants (blaKPC-3), overexpression of blaKPC and omp genes, and acquisition of metallo-beta-lactamase (MBL) genes.28,29 In vivo mutation D179Y of blaKPC-2 was firstly reported in ST307 K. pneumoniae isolated from a patient following only 12 days of CZA exposure in Puerto Rico.5 Subsequently, it occurred in the predominant CRKP clone ST11 K. pneumoniae in China, designated blaKPC-33 which associated with the emergence and recovery of CZA resistance.30,31 Herein, we noticed an in vivo mutation of K. pneumoniae blaKPC-2 into blaKPC-33 led to CZA drug resistance 14 days after CZA treatment, which restored the sensitivity of imipenem and meropenem and reduced MIC values of ertapenem. The result of conjugation assay further demonstrated the transmissibility of the plasmid carrying KPC-33. Of note, we also observed the elevated MIC for quinolone of the transconjugant strain, which indicated the co-conjugation of pFK8695-tetA plasmid carrying fluoroquinolone-resistant qnrS1 gene.
Acquisition of CZA resistance has been reported after CZA therapy in recent years. Resistance to CZA has been observed in strains with mutations in AmpC, blaKPC-2 and blaKPC-3, and the mutation point was mostly in Ω-loop in blaKPC genes.32,33 CZA resistance could appear as early as 6 days after administration, which poses major challenges to the clinical application and limits the therapeutic effect of CZA31. Fortunately, the resistant phenotype of KPC-33-producing K. pneumoniae could not be maintained without CZA selection pressure and would return to sensitive phenotype under the further selection pressure of imipenem.31 However, the emergence and prevalence of blaKPC-33 still need more attention and early detection of blaKPC-33 is important for the choice of medication.
Convergence of hypervirulence and carbapenem resistance in ST11 K. pneumoniae was mainly caused by acquisition of pLVPK-like virulence plasmid, which poses a consequential public health challenge.34 In FK8695, we identified a pLVPK-like plasmid pFK8695-rmpA with high similarity to the previously reported plasmid pVir-CR-hvKP-C789. The two plasmids were all from ST11-KL64 CRKP strains. However, pFK8695-rmpA misses siderophore synthesis operon iucABCD-iutA and iroBCDN, leaving only rmpA and a truncated iroN. It has been reported that aerobactin encoded by the iucABCD operon accounts for >90% of the siderophore activity and plays a crucial role in the growth and survival of hvKP in vivo and in vitro.35 However, our results showed that FK8578 still has a high fatality rate in G. mellonella infection model, which might contribute to the chromosome-encoded siderophore enterobactin and yersiniabactin or other virulence factors.
Furthermore, we noticed a mutation of the rmpA gene in CZA-resistant FK8695 compared to CZA-susceptible FK8578. The mutation was on the poly (G) region and might be mediated by DNA slip-strand synthesis of rmpA.36 Yu et al thought that mutation of rmpA systems contributes to the loss of the Hmv phenotype in the K. pneumoniae isolates positive for rmpA systems.37 This was in accordance with our result that FK8578 possessed Hmv phenotype but FK8695 was not. It has been reported that acquisition of colistin resistance in vivo or tigecycline resistance in vitro changed the Hmv phenotype.38,39 A recent study also showed that ST11 CR-hvKp underwent adaptive evolution during antibiotic treatment, including tigecycline resistance and virulence attenuation through mutations of ompK26 and acyltransferase (act) genes. Thus, we speculate that mutation of rmpA might be a compensatory mechanism of CR-hvKp counteracts the reduced fitness caused by the acquisition of CZA resistance. But further research is needed to confirm the relationship. In addition, whether this adaptive compensation will contribute to the further emergence of successful CZA-resistant clonal lineages also needs to be further evaluated.
Conclusion
In summary, this study describes the within-host acquisition of CZA resistance and virulence attenuation in ST11 CRKP during CZA treatment, and illuminate the mechanism of its resistance and virulence changes. Our study highlights the risk of mutations of blaKPC-2 gene in acquiring resistance to CZA and emphasizes the necessity to monitor the CZA resistance among CRKP.
Date Availability Statement
The datasets generated are available from the corresponding author on reasonable request.
Ethical Approval
This study was approved by the First Affiliated Hospital of Wenzhou Medical University Ethics Committee (Issuing No. 2022R140). Written informed consent for publication of their details was obtained from the patient.
Acknowledgments
We acknowledge the First Affiliated Hospital of Wenzhou Medical University for its help.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was funded by Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province (2022E10022) and the Planned Science and Technology Project of Wenzhou (no. Y2020974).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi:10.1093/infdis/jiw282
2. Han X, Shi Q, Mao Y, et al. Emergence of ceftazidime/avibactam and tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae due to in-host microevolution. Front Cell Infect Microbiol. 2021;11:757470. doi:10.3389/fcimb.2021.757470
3. Zasowski EJ, Rybak JM, Rybak MJ. The beta-lactams strike back: ceftazidime-avibactam. Pharmacotherapy. 2015;35(8):755–770. doi:10.1002/phar.1622
4. Shields RK, Potoski BA, Haidar G, et al. Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis. 2016;63(12):1615–1618. doi:10.1093/cid/ciw636
5. Giddins MJ, Macesic N, Annavajhala MK, et al. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother. 2018;62(3):1. doi:10.1128/AAC.02101-17
6. Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi:10.1016/j.gene.2004.05.008
7. Walker KA, Treat LP, Sepulveda VE, Miller VL. The small protein RmpD drives hypermucoviscosity in Klebsiella pneumoniae. mBio. 2020;11(5). doi:10.1128/mBio.01750-20
8. Nassif X, Fournier JM, Arondel J, Sansonetti PJ. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect Immun. 1989;57(2):546–552. doi:10.1128/iai.57.2.546-552.1989
9. Walker KA, Miner TA, Palacios M, et al. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio. 2019;10(2). doi:10.1128/mBio.00089-19
10. Ye M, Liao C, Shang M, et al. Reduced virulence and enhanced host adaption during antibiotics therapy: a story of a within-host carbapenem-resistant Klebsiella pneumoniae sequence type 11 evolution in a patient with a serious scrotal abscess”. mSystems. 2022;7:e0054522. doi:10.1128/msystems.00545-22
11. Xie M, Yang X, Xu Q, et al. Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun Biol. 2021;4(1):650. doi:10.1038/s42003-021-02148-4
12. Findlay J, Poirel L, Juhas M, Nordmann P. KPC-mediated resistance to ceftazidime-avibactam and collateral effects in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2021;65(9):e0089021. doi:10.1128/AAC.00890-21
13. Li D, Liao W, Huang HH, et al. Emergence of hypervirulent ceftazidime/avibactam-resistant Klebsiella pneumoniae isolates in a Chinese tertiary hospital. Infect Drug Resist. 2020;13:2673–2680. doi:10.2147/IDR.S257477
14. Zhang Y, Lin Y, Zhang X, et al. Combining colistin with furanone C-30 rescues colistin resistance of gram-negative bacteria in vitro and in vivo. Microbiol Spectr. 2021;9(3):e0123121. doi:10.1128/Spectrum.01231-21
15. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. In: CLSI Supplement M100.
16. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters; 2021. Available from: http://www.eucast.org.
17. Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705. doi:10.1084/jem.20030857
18. Palacios M, Miner TA, Frederick DR, et al. Identification of two regulators of virulence that are conserved in Klebsiella pneumoniae classical and hypervirulent strains. mBio. 2018;9(4). doi:10.1128/mBio.01443-18
19. Palacios M, Broberg CA, Walker KA, Miller VL. A serendipitous mutation reveals the severe virulence defect of a Klebsiella pneumoniae fepB mutant. mSphere. 2017;2(4). doi:10.1128/mSphere.00341-17
20. Chen L, Yu K, Chen L, et al. Synergistic activity and biofilm formation effect of colistin combined with PFK-158 against colistin-resistant gram-negative bacteria. Infect Drug Resist. 2021;14:2143–2154. doi:10.2147/IDR.S309912
21. Tsai CJ, Loh JM, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7(3):214–229. doi:10.1080/21505594.2015.1135289
22. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi:10.1101/gr.215087.116
23. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–6. doi:10.1093/nar/gkj014
24. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi:10.1093/bioinformatics/btu153
25. Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi:10.1093/jac/dkaa345
26. Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12(1):4188. doi:10.1038/s41467-021-24448-3
27. Zarate JO, Santos Lucero R, Padorno LO, Espiniella F, Apud A. Variantes para la demostracion histologica del Campylobacter pilorico con microscopia optica. [Variants for the histological demonstration of pyloric Campylobacter with optical microscopy]. Acta Gastroenterol Latinoam. 1987;16(4):233–240.
28. Sun L, Li H, Wang Q, Liu Y, Cao B. Increased gene expression and copy number of mutated blaKPC lead to high-level ceftazidime/avibactam resistance in Klebsiella pneumoniae. BMC Microbiol. 2021;21(1):230. doi:10.1186/s12866-021-02293-0
29. Haidar G, Clancy CJ, Shields RK, Hao B, Cheng S, Nguyen MH. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2017;61(5):1. doi:10.1128/AAC.02534-16
30. Shi Q, Yin D, Han R, et al. Emergence and recovery of ceftazidime-avibactam resistance in blaKPC-33-harboring Klebsiella pneumoniae sequence type 11 isolates in China. Clin Infect Dis. 2020;71(Suppl4):S436–S439. doi:10.1093/cid/ciaa1521
31. Wang C, Zhao J, Liu Z, et al. In vivo selection of imipenem resistance among ceftazidime-avibactam-resistant, imipenem-susceptible Klebsiella pneumoniae isolate with KPC-33 carbapenemase. Front Microbiol. 2021;12:727946. doi:10.3389/fmicb.2021.727946
32. Shirley M. Ceftazidime-avibactam: a review in the treatment of serious gram-negative bacterial infections. Drugs. 2018;78(6):675–692. doi:10.1007/s40265-018-0902-x
33. Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):124e1–124 e4. doi:10.1016/j.cmi.2019.08.020
34. Chen L, Kreiswirth BN. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis. 2018;18(1):2–3. doi:10.1016/S1473-3099(17)30517-0
35. Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. 2015;83(8):3325–3333. doi:10.1128/IAI.00430-15
36. Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. 2010;192(12):3144–3158. doi:10.1128/JB.00031-10
37. Yu WL, Lee MF, Tang HJ, Chang MC, Chuang YC. Low prevalence of rmpA and high tendency of rmpA mutation correspond to low virulence of extended spectrum beta-lactamase-producing Klebsiella pneumoniae isolates. Virulence. 2015;6(2):162–172. doi:10.1080/21505594.2015.1016703
38. Choi MJ, Ko KS. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother. 2015;59(11):6763–6773. doi:10.1128/AAC.00952-15
39. Park S, Lee H, Shin D, Ko KS. Change of hypermucoviscosity in the development of tigecycline resistance in hypervirulent Klebsiella pneumoniae sequence type 23 strains. Microorganisms. 2020;8(10):1. doi:10.3390/microorganisms8101562
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.