Back to Journals » OncoTargets and Therapy » Volume 13
Elevation in the Expression of circ_0079586 Predicts Poor Prognosis and Accelerates Progression in Glioma via Interactions with the miR-183-5p/MDM4 Signaling Pathway
Authors Chen J , Liu T, Wang H, Wang Z, Lv Y, Zhao Y, Yang N, Yuan X
Received 15 October 2019
Accepted for publication 27 March 2020
Published 8 June 2020 Volume 2020:13 Pages 5135—5143
DOI https://doi.org/10.2147/OTT.S234758
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Jingyu Chen,1 Tianyi Liu,2 Hui Wang,3 Zhipeng Wang,1 Yanju Lv,4 Yuying Zhao,1 Ning Yang,1 Xueli Yuan1
1Department of Oncology, The Fourth Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province 150001, People’s Republic of China; 2Department of Pathology, The Second Hospital of Harbin Medical University, Harbin, Heilongjiang Province 150086, People’s Republic of China; 3Department of Oncology, Shandong Cancer Hospital Affiliated to Shandong University, Jinan, Shandong Province 250117, People’s Republic of China; 4Department of Oncology, The Second Hospital of Harbin Medical University, Harbin, Heilongjiang Province 150086, People’s Republic of China
Correspondence: Xueli Yuan
Department of Oncology, The Fourth Affiliated Hospital of Harbin Medical University, No. 37, YiYuan Street, NanGang District, Harbin 150001, People’s Republic of China
Tel +86-451-85939760
Email [email protected]
Purpose: Glioma (GM) usually presents with an aggressive behavior and has a poor survival outcome. The abnormal expression of circular RNAs (circRNAs) has already been detected in GM, and circ_0079586 was found to have an increased expression in GM tumors.
Patients and Methods: We assessed the differences in the expression of circ_0079586 in GM tissues (N=60) and cell lines (N=5) using qRT-PCR. The clinical value of circ_0079586 was measured by Fisher’s exact test and Kaplan–Meier and Cox regression analyses. Circ_0079586 siRNA and vector were transfected into LN229 and U251 cells, respectively, and the transfection was verified by qRT-PCR. Cell growth was evaluated by cell counting kit-8 (CCK-8). Cell apoptosis was measured using flow cytometric assay. Cell metastatic properties were measured by wound scratch and transwell experiments. Subcellular fractionation was used to identify the location of circ_0079586. Dual-luciferase reporter test was utilized to confirm the interaction between miR-183-5p and circ_0079586/MDM4 3ʹ-UTR.
Results: The expression of circ_0079586 was elevated in GM samples and cells and correlated with the clinical severity and unfavorable prognosis of the patients. The elevated expression of circ_0079586 led to an increase in cell growth, migration and invasion but inhibited apoptosis in U251 cells, whereas its down-regulation reversed these effects in the LN229 cells. Mechanistically, we found circ_0079586 to be primarily located in the cytoplasm of GM cells. Furthermore, circ_0079586 could act as a sponge for miR-183-5p and elevate MDM4 expression at the posttranscriptional level.
Conclusion: In summary, circ_0079586 was found to be up-regulated in GM that increased the proliferation, migration and invasion in GM cells via interaction with the miR-183-5p/MDM4 axis. We anticipate that our study would provide newer insights into the mechanism and treatment of GM.
Keywords: glioma, circular RNA, circ_0079586, miR-183-5p, MDM4 protein
Introduction
Glioma (GM), the most commonly found tumor in the brain, has shown an increasing trend in its incidence and mortality rate, making it a deadly malignancy worldwide.1,2 Although improved diagnostic and therapeutic approaches have improved patient survival, the prognosis continues to remain poor.3 Currently, the biomarkers for prognosis of GM are mainly based on the clinical and pathological stage of the tumor.3 However, significantly different progression and outcomes could be observed in patients that presented with same clinical symptoms and were treated with similar therapeutic regimens. Therefore, novel prognostic factors need to be identified on priority basis for the GM patients.
Circular RNAs (circRNAs) are a class of non-coding RNA molecules with closed circular structures and lack the ability to encode proteins.4,5 These RNAs take an important effect in generation, proliferation and metastasis of tumor cells by modulating the oncogenic and tumor suppressor pathways.6 Further, numerous studies have shown that the abnormal regulation of circRNAs in a variety of tumors may affect cell proliferation and apoptosis, together with their role in tumorigenesis and drug resistance.7–10 As a tumorigenic circRNA, circ_0030235 was found to be overexpressed in pancreatic ductal adenocarcinoma and promoted cell proliferation and metastasis by targeting miR-1253 and miR-1294.11 In breast cancer, hsa_circ_0004771 contributed to cell oncogenesis by modulating the miR-653/ZEB2 signaling pathway.12 However, the expression, mechanism, and clinical significance of circRNAs in GM remain to be fully investigated.
Wang screening high-throughput sequencing of circRNA on GM specimens and their corresponding non-cancer tissues, and found circ_0079586 to be up-regulated in the GM tissues.13 Circ_0079586 is located at chr7: 23353140–23383472 and is generated by splicing of IGF2BP3. This circRNA is 586 base pairs (bp) in length. Here, our analysis has identified the circ_0079586/miR-183-5p/MDM4 axis has a certain impact on cell growth, apoptosis, migration and metastasis in GM, and may serve as a newer biomarker for prognosis of the patients.
Materials and Methods
Ethics Statement
According to the ethical principle, which is proclaimed in the Declaration of Helsinki, ethical approval was acquired under the discussion debated by the ethical board of the Fourth Affiliated Hospital of Harbin Medical University (ethical approval code 2020-CILLSC-02).
Experimental Specimens and Cell Culture
Tissue samples were acquired from all the patients with informed consent and approval for the study was obtained from the Ethics Committee of the Affiliated Hospital of Harbin Medical University. A total of sixty paired fresh tumor and corresponding noncancerous tissue specimens were obtained and stored at −80°C.
Human GM cell lines (U87MG, U251, U118 and LN229H4) and NHA cells were purchased from the Chinese Academy of Sciences (Shanghai). All the cell lines were cultured in DMEM/RPMI-1640 containing 10% FBS at 37°C and in an atmosphere containing 5% CO2.
qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). MicroRNA levels were detected using the TaqMan MicroRNA Assay. GAPDH and U6 small nuclear RNA (U6 snRNA) were used as the reference RNAs. Relative RNA expression was measured using the SYBR Green System. The 2−ΔΔCt method was used to determine the relative RNA levels.
Cell Transfection
The miR-183-5p mimics and inhibitor, siRNAs targeting circ_0079586 and MDM4, and the negative controls for each were purchased from Genepharma (Shanghai, China). The pcDNA-3.1-circ_0079586 mini vector and pcDNA.3.1-MDM4 were purchased from the GeneSeed (Guangzhou, China). Transfection assays were performed using Lipofectamine 3000 (Invitrogen, USA) as per the directions of the manufacturer.
Cell Counting Kit-8 (CCK-8)
CCK-8 assay (Bimake, Houston, USA) was performed to assess the effect of circ_0079586 on LN229 and U251 cells growth. 2×103 cells/well were plated into 96-well plates. Absorbance was measured at 450 nm using a microplate every 24 hours to assess cell proliferation after addition of 10 μL of the CCK-8 reagent and incubated at 37°C.
Cell Apoptosis Assay
Apoptosis assay was used Annexin V-FITC and propidium iodide (PI; BD Biosciences, USA) for double staining. Briefly, collect and resuspend the cells in Binding Buffer, add AnnexinV-FITC and PI, incubate for 10 minutes in the dark at room temperature, and then subjected to flow cytometric analysis.
Wound Scratch Test
Each well was overspread with 5 x 105 cells. Scratch the vertical line of the monolayer cells with a 200μL pipette tip. After capturing images at 0h and 36h, the images were processed using the Image J software.
Transwell Assay
In the cell migration and invasion test, LN229 and U251 cells were placed in 24-well transwell chambers, and cell migration assays were performed using pre-coated Matrigel, and cell invasion assay was performed without Matrigel. Fill the lower compartment with complete medium. After 24h of culture, non-migrating/invasive cells were removed with a cotton swab, and the migrating/invasive cells were fixed with methanol and then stained with crystal violet. Count and capture images of stained cells in each well.
Western Blotting Assay
Proteins were isolated by RIPA buffer, quantified, 30µg protein was added to each lane of SDS-PAGE gel for electrophoretic separation, and the protein was transferred onto PVDF membrane. The membranes were blocked and then exposed to antibodies against GAPDH or MDM4 (Abcam, Cambridge, MA, USA). Further, the blots were incubated with the secondary antibody (Zhongshanjinqiao, China). RapidStep ECL Reagent (Millipore, Billerica, MA, USA) was used to analyze the intensity of each protein band.
Subcellular Fractionation
Cytoplasmic and nuclear RNAs were isolated with the PARIS Kit (Life Technologies, USA) and quantified by qRT-PCR with the internal references of GAPDH and U6, respectively.
Luciferase Reporter Assay
To detect whether miR-183-5p could bind the 3ʹ-UTR of MDM4, we cloned the 3ʹ-UTR amplification product of MDM4 downstream of the firefly luciferase gene in the pGL3 vector (Promega). We named the vector as wild-type (WT). GeneTailor Site-Directed Mutagenesis System (Invitrogen, USA) was used to perform site-directed mutagenesis of the miR-183-5p binding sites in the 3ʹUTR of MDM4. We named this vector as mutant (MUT). Cells were co-transfected with the vector and the miR-183-5p mimics or negative control. Similarly, the mutant seed or wild-type region of circ_0079586 carrying the putative target site for the miR-183-5p was cloned into the pGL3 vector. Cells were co-transfected with the MUT or WT circ_0079586 vector and the miR-183-5p mimic or negative control. Thirty-six hours post-transfection, the Dual-Luciferase Reporter Assay System (Promega) was used to assess the extent of interaction in each set.
Data Analysis
Data were analyzed using the SPSS software, version 20.0 (Chicago, IL, USA). Two-tailed t-test of one-way ANOVA was used to compare groups. Spearman correlation was used to determine the correlation between the expression of circ_0079586 and miR-183-5p/MDM4. The clinical significance of circ_0079586 was evaluated by the Fisher’s exact test, Kaplan–Meier survival curve, and Cox regression analysis. P < 0.05 represents statistically significant values.
Results
Circ_0079586 Is Overexpressed in GM Tissues and Cells and Associates with Unfavorable Prognosis
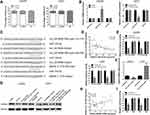
First, total tissue RNA was isolated from clinical GM specimens and matched normal brain samples (N = 60). The qRT-PCR analysis identified an increased expression of circ_0079586 in GM than in normal brain tissues (Figure 1A). Additionally, it was found to be up-regulated in the GM cell lines than in the NHA cells (Figure 1B). And then, we explored the clinical significance of the expression of circ_0079586 and separated the patients into two groups on the basis of the median value of circ_0079586, namely the high expression group or the low expression group. We also examined the correlation between the patient’s clinical features and circ_0079586 expression. Our analysis suggested that the circ_0079586 expression was significantly elevated in the GM samples (WHO tumor grade III and IV) than in the WHO tumor grade I and II samples (P = 0.001). While the expression of circ_0079586 correlated with tumor size (P = 0.037), it did not correlate with factors such as age, gender, and tumor location (P > 0.05) (Table 1). Interestingly, high expression of circ_0079586 correlated with poor overall survival in GM patients (Figure 1C). Furthermore, while the univariate analysis indicated WHO grade (P = 0.032) and circ_0079586 expression (P = 0.010) as prognostic indicators for GM patients, the multivariate analysis failed to demonstrate the prognostic role of circ_0079586 (P > 0.05) (Table 2). Taken together, our analysis indicates that the circ_0079586 is over-expressed in GM, and correlates with tumor size and poor survival outcome.
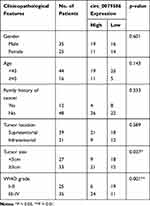 |
Table 1 Correlation Between circ_0079586 Expression and Clinicopathological Parameters of Glioma Patients |
 |
Table 2 Univariate and Multivariate Analysis of Prognostic Factors for Overall Survival in Glioma Patients |
Circ_0079586 Contributes to Progression in GM Cells
For the highest expression of circ_0079586, LN229 cell line was chosen for the knockdown study (Figure 1B). Two siRNAs targeting circ_0079586 were tested for decreasing circ_0079586 levels in LN229 cells, of which si-circ_0079586-1 showed the highest efficiency and was chosen for subsequent experiments (Figure 2A). Further, the U251 cell showed the lowest expression of circ_0079586 and was chosen for the overexpression study (Figure 2B). The analysis of the CCK-8 assay indicated that the cell viability was significantly reduced upon knockdown of circ_0079586 in LN229 cells, whereas, its ectopic expression increased cell growth in U251 cells (Figure 2C). Next, transfection of si-circ_0079586 increased the rate of cell apoptosis in LN229 cells, and its over-expression reduced apoptosis of U251 cells (Figure 2D). The knockdown of circ_0079586 suppressed the cell migration of LN229 cells, while migration increased with its over-expression in U251 cells (Figure 2E). Additionally, similar results were obtained with the cell invasion assay upon knockdown and over-expression of circ_0079586 (Figure 2F). Taken together, the above results show that circ_0079586 has important function in modulating the cell growth, apoptosis, cell migration and cell invasion phenotype of GM cells.
Circ_0079586 Regulates the Expression of MDM4 by Sponging miR-183-5p
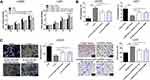
As described earlier, the subcellular location of circRNA determines its specific functions.6 Our analysis of the subcellular fractions indicated that circ_0079586 was enriched in the cytoplasm of LN229 (79.5%) and U251 (78.6%) cells (Figure 3A). Based on this observation, we hypothesized that circ_0079586 may regulate the posttranscriptional activity in GM cells. We analyzed the circular RNA interactome that predicted 26 candidate miRNAs, while the circBANK database identified 70 miRNAs as possible candidates to bind circ_0079586. An overlap of these datasets identified three miRNAs (miR-127-5p, miR-183-5p and miR-433) to be common. We validated the expression of these miRNAs using qRT-PCR, which indicated only miR-183-5p to be negatively regulated by circ_0079586 in GM cells (Figure 3B). Further, our bioinformatics analysis predicted circ_0079586 to harbor conserved binding sites for miR-183-5p with a high score (312–318; 539–545, Figure 3C). Additionally, our analysis suggested an inverse expression of circ_0079586 and miR-183-5p (Figure 3D). Subsequently, luciferase reporter assay was carried out in the LN229 and U251 cells to confirm the bioinformatics prediction. The results indicated that overexpression of miR-183-5p significantly decreased the luciferase signal intensity in WT cells (Figure 3E). To understand the biological interactions of miR-183-5p in GM, the putative targets were searched using TargetScan7.2. As shown in Figure 3C, miR-183-5p also had complementary binding sites in the 3ʹUTR of MDM4. Moreover, the transcript levels of MDM4 were decreased upon transfection of miR-183-5p mimics in LN229 and U251 cells than with miR-NC, whereas, the miR-183-5p inhibitor reversed the effect (Figure 3F). Next, the Western blot analysis indicated that the relative expression of MDM4 was significantly down-regulated in LN229 cells transfected with si-circ_0079586, whereas, inhibition of miR-183-5p or over-expression of MDM4 rescued the expression of MDM4 (Figure 3G). Further, in U251 cells, either miR-183-5p mimics or si-MDM4 partially decreased the expression of circ_0079586 (Figure 3G). Additionally, we identified a positive correlation between the expression of circ_0079586 and MDM4 in the GM tissue samples (Figure 3H). Moreover, the luciferase reporter test indicated that the luciferase activity of MDM4 3ʹ-UTR WT was significantly reduced, but not MDM4 3ʹUTR MUT, in the LN229 and U251 cells transfected with miR-183-5p mimics (Figure 3I). Taken together, these results suggest circ_0079586 to regulate MDM4 by sponging the activity of miR-183-5p.
Circ_0079586 Increases GM Cell Oncogenesis by Regulating the miR-183-5p/MDM4 Pathway
To elucidate the relationship between circ_0079586, miR-183-5p and MDM4 in GM, si-circ_0079586 and miR-183-5p inhibitor were co-transfected into the LN229 cells. As shown in Figure 4A–C, silencing the expression of circ_0079586 inhibited cell proliferation and invasion, and boosted cell apoptosis, whereas inhibition of miR-183-5p or MDM4 vector rescued these phenotype. On the contrary, the oncogenic properties induced by circ_0079586 vector were partly inhibited by miR-183-5p mimics or si-MDM4 (Figure 4A–C).
Discussion
CircRNAs have been reported to serve either as oncogenes or tumor suppressors during progression of several cancers, including GM.14 The circ_0079593 was found to be up-regulated, and its overexpression elevated cell proliferation and restrained cell apoptosis by targeting miR-182 and miR-433 in GM cells.15 Hsa_circ_0014359 was shown to facilitate GM progression via the PI3K signaling pathway by sponging miR-153.16 Further, Wang et al used circRNA microarrays to screen circRNAs differentially expressed in GM neoplasm and non-neoplasm tissues. Wang et al screened the differentially expressed circRNAs in GM tumor and non-tumor tissues using the circRNA microarray.13 From their list of up-regulated circRNAs, we chose circ_0079586 to investigate its role in GM. circ_0079586 is located on chr7: 23353140–23383472 and is back-spliced from the IGF2BP3 gene to generate a 586bp transcript.
Here, we observed up-regulation of circ_0079586 in four cell lines and sixty cases of GM tissues. Moreover, the over-expression of circ_0079586 correlated with clinicopathological factors such as tumor size and advanced WHO grade tumors in GM patients. Furthermore, Kaplan–Meier survival analysis showed that the overall survival rate of GM patients with increased circ_0079586 expression was lower than that of patients with lower circ_0079586 levels. However, the multivariate analysis failed to demonstrate the independent prognostic role of circ_0079586, possibly due to the smaller sample size, and a larger patient cohort should allow making inference the Cox regression analysis. Next, our in vitro functional experiments indicated that circ_0079586 could promote cell growth, migration and invasion in GM cell lines. Additionally, the oncogenic functions of circ_0079586 can be partially attributed to the inhibition of cell apoptosis, together suggesting its role in occurrence and progression of GM.
Studies indicate that circRNAs play a part of competing endogenous RNAs (ceRNAs) for sponge miRNAs, thereby affecting the function of target mRNA expression.12–16 Interestingly, we found circ_0079586 to be localized in the cytoplasm, and to act as a miR-sponge for miR-183-5p, thus supporting its potential role as a ceRNA. Several studies have shown that the expression level of miR-183-5p is low in many cancers.17–19 Downstream targets of miR-183-5p, such as Erbin, PIK3CA, Ezrin, and so on, have been identified in various cancer types.17–19 Consistent with these reports, our analysis identified low expression of miR-183-5p in GM tissues. Using TargetScan and luciferase reporter assay, we validated miR-183-5p to bind the 3ʹUTR of MDM4 in GM cells. Further, we confirmed their association by silencing and over-expressing miR-183-5p that led to inversely related expression pattern of MDM4 in these cells. MDM4 is known to inhibit the transcriptional activity of p53 by binding to the E3 ubiquitin ligase of MDM2 and promoting p53 ubiquitination and proteasomal degradation.20,21 Multiple studies have demonstrated MDM4 to be regulated at the posttranscriptional level and play an oncogenic role in cancers of the lung (non-small cell lung cancer), the colon, and the pancreas.22–24 Furthermore, MDM4 has been shown to increase the hazard of GM susceptivity in the Han Chinese population.25 Additionally, the study by Xie et al identified let-7 to bind MDM4 and contribute to the DNA damage response in GM.26 Thus, these reports together with our analysis implicate MDM4, and its regulators and targets to be involved in tumor formation and progression in GM.
Conclusion
In conclusion, our analysis demonstrates the circ_0079586/miR-183-5p/MDM4 pathway to possibly regulate GM cell progression. Moreover, we anticipate this newer regulatory network to provide novel biomarkers for diagnosis and prognosis in patients with GM.
Disclosure
All authors have declared no conflicts of interest in this work.
References
1. Malzkorn B, Reifenberger G. Practical implications of integrated glioma classification according to the World Health Organization classification of tumors of the central nervous system 2016. Curr Opin Oncol. 2016;28:494–501.
2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi:10.1007/s00401-016-1545-1
3. Mizumoto M, Yamamoto T, Ishikawa E, et al. Proton beam therapy with concurrent chemotherapy for glioblastoma multiforme: comparison of nimustine hydrochloride and temozolomide. J Neuro Oncol. 2016;130:165–170. doi:10.1007/s11060-016-2228-4
4. Memczak S, Jens A, Elefsinioti M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:331–338. doi:10.1038/nature11928
5. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi:10.1038/nbt.2890
6. Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
7. Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455–461. doi:10.1016/j.bbrc.2018.01.077
8. Mao Y, Zhang L, Li Y. circEIF4G2 modulates the malignant features of cervical cancer via the miR‑218/HOXA1 pathway. Mol Med Rep. 2019;19:3714–3722. doi:10.3892/mmr.2019.10032
9. Ma HB, Yao YN, Yu JJ, et al. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10:592–604.
10. Xu Y, Yao Y, Liu Y, et al. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma. Aging (Albany NY). 2019. doi:10.18632/aging.101872
11. Xu Y, Yao Y, Gao P, et al. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509:138–142. doi:10.1016/j.bbrc.2018.12.088
12. Xie R, Tang J, Zhu X, et al. Silencing of hsa_circ_0004771 inhibits proliferation and induces apoptosis in breast cancer through activation of miR-653 by targeting ZEB2 signaling pathway. Biosci Rep. 2019;39. doi:10.1042/BSR20181919
13. Wang R, Zhang S, Chen X, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17:166. doi:10.1186/s12943-018-0911-0
14. Liu J, Zhao K, Huang N, et al. Circular RNAs and human glioma. Cancer Biol Med. 2019;16:11–23. doi:10.20892/j.issn.2095-3941.2018.0425
15. Qu Y, Zhu J, Liu J, et al. Circular RNA circ_0079593 indicates a poor prognosis and facilitates cell growth and invasion by sponging miR-182 and miR-433 in glioma. J Cell Biochem. 2019;120:18005–18013. doi:10.1002/jcb.29103
16. Shi F, Shi Z, Zhao Y, et al. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem Biophys Res Commun. 2019;510:614–620. doi:10.1016/j.bbrc.2019.02.019
17. Zheng Z, Zheng X, Zhu Y, et al. miR-183-5p inhibits occurrence and progression of acute myeloid leukemia via Targeting Erbin. Mol Ther. 2019;27:542–558. doi:10.1016/j.ymthe.2019.01.016
18. Meng F, Zhang L. miR-183-5p functions as a tumor suppressor in lung cancer through PIK3CA inhibition. Exp Cell Res. 2019;374:315–322. doi:10.1016/j.yexcr.2018.12.003
19. Yan H, Sun BM, Zhang YY, et al. Upregulation of miR-183-5p is responsible for the promotion of apoptosis and inhibition of the epithelial-mesenchymal transition, proliferation, invasion and migration of human endometrial cancer cells by downregulating Ezrin, Int. J Mol Med (Berl). 2018;42:2469–2480.
20. Gerhart SV, Kellner WA, Thompson C, et al. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci Rep. 2018;8:9711. doi:10.1038/s41598-018-28002-y
21. Miranda PJ, Buckley D, Raghu D, et al. MDM4 is a rational target for treating breast cancers with mutant p53. J Pathol. 2017;241:661–670. doi:10.1002/path.4877
22. Chen W, Cai G, Liao Z, et al. miRNA-766 induces apoptosis of human colon cancer cells through the p53/Bax signaling pathway by MDM4. Exp Ther Med. 2019;17:4100–4108. doi:10.3892/etm.2019.7436
23. Yan H, Chen X, Li Y, et al. MiR-1205 functions as a tumor suppressor by disconnecting the synergy between KRAS and MDM4/E2F1 in non-small cell lung cancer. Am J Cancer Res. 2019;9:312–329.
24. Han H, Wang L, Xu J, et al. miR-128 induces pancreas cancer cell apoptosis by targeting MDM4. Exp Ther Med. 2018;15:5017–5022. doi:10.3892/etm.2018.6047
25. Sun P, Yan F, Fang W, et al. MDM4 contributes to the increased risk of glioma susceptibility in Han Chinese population. Sci Rep. 2018;8:11093. doi:10.1038/s41598-018-29468-6
26. Xie C, Chen W, Zhang M, et al. MDM4 regulation by the let-7 miRNA family in the DNA damage response of glioma cells. FEBS Lett. 2015;589:1958–1965. doi:10.1016/j.febslet.2015.05.030
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.