Back to Journals » Cancer Management and Research » Volume 11
EIF3B is associated with poor outcomes in gastric cancer patients and promotes cancer progression via the PI3K/AKT/mTOR signaling pathway
Authors Wang L , Wen X, Luan F, Fu T, Gao C, Du H, Guo T, Han J, Huangfu L, Cheng X , Ji J
Received 8 March 2019
Accepted for publication 30 May 2019
Published 21 August 2019 Volume 2019:11 Pages 7877—7891
DOI https://doi.org/10.2147/CMAR.S207834
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Lin Wang,1,2 Xianzi Wen,1 Fengming Luan,1 Tao Fu,2 Chao Gao,2 Hong Du,1 Ting Guo,1 Jing Han,1 Longtao Huangfu,1 Xiaojing Cheng,1 Jiafu Ji1,2
1Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Division of Gastrointestinal Cancer Translational Research Laboratory, Peking University Cancer Hospital & Institute, Beijing, People’s Republic of China; 2Department of Gastrointestinal Surgery, Peking University Cancer Hospital & Institute, Beijing, People’s Republic of China
Correspondence: Xiaojing Cheng
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Division of Gastrointestinal Cancer Translational Research Laboratory, Peking University Cancer Hospital & Institute, Beijing 100142, People’s Republic of China
Tel +86 108 819 6760
Email [email protected]
Jiafu Ji
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Gastrointestinal Surgery, Peking University Cancer Hospital & Institute, Beijing 100142, People’s Republic of China
Tel +86 108 819 6048
Email [email protected]
Purpose: Eukaryotic translation initiation factor (EIF) plays a vital role in protein synthesis. EIF3B is a core subunit of the EIF3 family, and is overexpressed in many tumors. EIF3B is associated with an unfavorable prognosis, as well as the genesis and development of tumors. However, the potential role of EIF3B in gastric cancer (GC) remains unknown. In the current study, we explored the clinical significance and the possible mechanism of EIF3B in the progression of GC.
Methods: EIF3B expression was analyzed in 78 GC tissue samples through quantitative PCR and in 94 GC tissue samples through immunohistochemistry (IHC) staining. The correlation between EIF3B and clinicopathological features was analyzed in GC tissues. The role of EIF3B in GC progression was investigated through in vitro and in vivo assays.
Results: EIF3B expression was upregulated in GC tissues (73.4%, IHC). High expression of EIF3B was significantly correlated with the depth of tumor invasion, lymph node metastasis and TNM stage (P=0.000, 0.000 and 0.000, respectively). Multivariate analysis indicated that GC patients with high EIF3B expression suffered a poorer 5-year survival. EIF3B promoted GC cell proliferation and was strongly associated with proliferating cell nuclear antigen (PCNA) expression in GC samples (P=0.009). It also enhanced tumor cell migration and invasion, which were affected through epithelial-mesenchymal transition (EMT) and the Stat3 signaling pathway. Knockdown of EIF3B in GC cells suppressed the growth of xenograft tumors and lung metastatic colonization in vivo. Furthermore, gene set enrichment analysis (GSEA) and Western blot results demonstrated that EIF3B activated the PI3K/AKT/mTOR signaling pathway.
Conclusion: Our results suggest that EIF3B plays an oncogenic role in GC progression and serves as an independent prognostic factor for GC patients.
Keywords: EIF3B, gastric cancer, prognosis, PI3K/AKT/mTOR pathway
Introduction
Gastric cancer (GC) is the fifth most common malignancy and the third leading cause of cancer-related death worldwide.1 Treatment of GC involves surgery, radiotherapy, chemotherapy, immunotherapy and molecularly targeted therapy, of which the preferred treatment option is surgery.2,3 In spite of considerable advances in therapeutic methods, GC prognosis has not been substantially improved, partly because of late stage diagnosis and a lack of reliable biomarkers for early detection.4,5 Tumor metastasis and recurrence are considered to be the most significant determinants for treatment failure and mortality.6 Therefore, exploring new prognostic molecular biomarkers and therapeutic targets is necessary, as well as clarifying underlying mechanisms.
Translation is an early step in protein synthesis, and represents an important point at which to exert regulatory control.7 There are various types of translation initiation factors which participate in the initial process of translation,8 and among them eukaryotic translation initiation factor (EIF) plays a pivotal role in forming the initiation complex that participates in assembling the 80S-mRNA-Met-tRNA complex to initiate protein synthesis.9,10 As a central member of the EIF family, EIF3 is the largest and most complex factor and is composed of 13 non-identical protein subunits (a–m), which are involved in almost the entire process of translation initiation regulation.8,11 The EIF3B subunit, also known as EIF3p110, EIF3p116 and hPrt1,12 is widely regarded as a critical scaffold protein in the EIF3 complex, which plays an essential role in translation regulation and cell growth.13 A previous study has confirmed that ectopic expression of EIF3B causes malignant transformation of immortal fibroblast cells (NIH3T3).14 Moreover, high expression of EIF3B was observed in several types of cancer and is associated with poor prognosis,12 such as clear cell renal cell carcinoma,15 esophageal squamous cell carcinoma,16 and bladder cancer.17 However, little is known about the relevance of EIF3B in GC.
In the present study, we first assessed the clinical significance of EIF3B in GC patients. In vitro results demonstrated that EIF3B promoted GC cell proliferation, migration and invasion. Further biological function analysis indicates that EMT process and the PI3K/AKT/mTOR axis was participated in this study. We also investigated the effect of EIF3B in tumor growth and lung metastasis using a xenograft mouse model. Our results suggest that EIF3B plays a key role in GC progression and predicted patient outcome.
Materials and methods
Clinical specimen collection
mRNA samples for quantitative PCR (qPCR) were isolated from 78 GC tissue samples. Tissue microarray (TMA) sets for IHC included 94 GC tissue samples. All specimens collected from GC patients who underwent radical gastrectomy between November 2005 and December 2010 were obtained from the Biobank of Peking University Cancer Hospital. A total of 15 pairs of fresh GC and adjacent normal tissues frozen in liquid nitrogen immediately after dissection were collected in November 2017 from the Department of Gastrointestinal Surgery of Peking University Cancer Hospital. None of the patients received chemotherapy or radiation therapy prior to surgery.
Clinicopathological and follow-up information was obtained from patient data. GC stage was classified based on the 2017 TNM classification recommended by the American Joint Committee on Cancer (AJCC 8th edition).18 Overall survival was defined as the time elapsed from the date of surgery to death or to the last follow-up, and disease-free survival was defined as the time elapsed from the date of surgery to the discovery of local relapse or distant metastasis. The survival of patients was tracked until December 2015. All patients involved in this study have written informed consent, in compliance with Declaration of Helsinki. The Ethics Committee of the Peking University Beijing Cancer Hospital have approved this clinical specimen study.
RNA extraction and qPCR analysis of EIF3B expression
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and cDNA was synthesized using M-MLV reverse transcriptase (Promega, Madison, WI, USA) in accordance with the manufacturer’s instructions. The expression level of EIF3B was determined using an ABI 7500 Fast instrument and a Power SYBR Green Master Mix (ThermoFisher Scientific Inc, Waltham, MA). Each sample was tested in triplicate, and the ∆∆CT method was used to analyze the fold change in EIF3B gene expression. The EIF3B primer sequences were: forward primer 5ʹ-GAC CGC ACT TCC ATA TTC TGG-3ʹ, reverse primer 5ʹ-GCA CAT ACG TCT CTG TCC ATC T −3ʹ. The internal standard GAPDH primer sequences were: forward primer 5ʹ-GAA GGT GAA GGT CGG AGT-3ʹ, reverse primer 5ʹ-GAA GAT GGT GAT GGG ATT TC −3ʹ.
Immunohistochemistry
Immunohistochemistry (IHC) for EIF3B was performed on sections of TMAs using an anti-EIF3B antibody (1:100) (ab133601, Abcam, UK). As a negative control, the sections were processed using the same protocol except that they were incubated in blocking solution without the primary antibody. EIF3B expression was evaluated based on the area of positive cytoplasmic staining. The intensity of the staining was categorized from negative (-) to low (+), intermediate (++), and high (+++). The expression of EIF3B was assessed independently by two experienced pathologists (B. D. and Z. L.), who were blind to the patients’ clinical outcomes. There was a high level of consistency between the two pathologists, and a consensus was reached after joint review in the few discrepant cases (<5%). Finally, the staining levels of EIF3B expression were denoted as: - and +, low expression; ++ and +++, high expression.
Cell lines
Human GC cells AGS, N87, and HEK293FT cell lines were purchased from ATCC (Manassas, VA, USA). The MKN28 cell line was obtained from the Health Science Research Resources Bank (Tokyo, Japan). GES1, BGC823, MGC803 and SGC7901 cell lines were obtained from the Cell Research Institute (Shanghai, China). The cells were cultured in RPMI-1640 medium (GIBCO, Carlsbad, NY, USA), supplemented with 10% (v/v) fetal bovine serum (GIBCO, NY, USA) at 37 °C in a humidified 5% CO2 atmosphere.
Plasmid construct and transfection
Lentivirus was produced through the co-transfection of HEK293FT cells with short hairpin RNA constructs targeting EIF3B (shControl, shEIF3B-1: 5ʹ-GGA AGC AGA UGG AAU CGA UTT-3ʹ, shEIF3B-2: 5ʹ-GCA CCU ACC UGG CUA CCU UTT-3ʹ; Shanghai Genephama Co., Ltd, China) and a lentiviral packaging mix (Invitrogen, Carlsbad, USA) in accordance with the manufacturer’s instructions. Lentivirus-containing supernatant was harvested at 48 h post-transfection, centrifuged, and stored at −80 °C. For viral transductions, 1 ml of the scrambled shControl or shEIF3B lentiviruses was incubated with BGC823 and SGC7901 cells for 48 h at 37 °C in a humidified cell culture incubator. Stable EIF3B knockdown GC cells were selected using puromycin (1 μg/ml) for 14 d, and the EIF3B protein expression level was identified through Western blot.
Protein extraction and Western blotting analysis
Total proteins were prepared from fresh-frozen tissues, and proteins were isolated from GC cell lines when they reached 80% confluency. The cells were washed twice using 1×phosphate buffer solution (1×PBS) before being completely lysed in pre-chilled RIPA lysis buffer containing a protease inhibitor cocktail for 30 min. The samples were then centrifuged at 15,000× g for 20 min.
Equal amounts of total protein extracts were separated through 8%, 10% or 12% SDS polyacrylamide gel electrophoresis, based on the molecular weight of the target protein, and transferred onto a 0.22 μm nitrocellulose filter membrane. The membrane was blocked for 1 h at room temperature using blocking buffer containing 5% nonfat dry milk, and then the membrane was incubated with primary antibodies diluted in blocking buffer at 4 °C overnight. After three washes in tris-buffered saline tween-20 (10 min each), the membrane was incubated with an appropriate secondary antibody, such as goat-anti-rabbit IgG (dilution 1:5000) or horse-anti-mouse IgG (dilution 1:5000), washed and developed using an enhanced chemiluminescence detection system (Amersham, GE Healthcare, Chicago, IL, USA).
The primary antibodies are as follows: E-cadherin (CST, #3195), N-cadherin (CST, #4061), Bax (CST, #2774), P53 (CST, #9282), Snail (CST, #3879), Slug (CST, #9585), Vimentin (CST, #5741), P-P53 (CST, #2521), Stat3 (CST, #12640), P-Stat3 (CST, #52075), Bcl2 (CST, #15071), P-Bcl2 (CST, #2827), PI3K (CST, #4249), Akt (CST, #4685), P-Akt (CST, #4060), mTOR (CST, #2972), P-mTOR (CST, #5536), GAPDH (CST, #5174) and EIF3B (Abcam, #ab133601). These antibodies were diluted at 1:1000.
Cell proliferation assay
GC cells were seeded in a RTCA E-Plate 16 (RTCA, xCELLigence Roche, Penzberg, Germany) at 2×103 cells per well. The baseline was measured and 10% fetal bovine serum was added to the lower chamber, in accordance with the xCELLigene manufacturer’s instructions. Plates were incubated for 30 min at room temperature before starting the measurements. The cell index was measured in a time-resolved manner (every 10 min during 96 h). Three independent experiments were performed.
Wound healing assay
Targeted cells were seeded into a 6-well plate. When cells grew to 80–90% confluency, cell monolayers were scraped using a sterile micropipette tip. The wounded monolayers were washed using 1×PBS to remove cell debris. The distance between the two edges of the wound was calculated at three different positions. A total of 36 h later, the distance between the two edges were measured again. All experiments were conducted three times in triplicate.
Boyden chamber invasion assay
For Boyden chamber-based invasion assays, 5×104 cells were loaded into an insert, provided with serum-free medium, and allowed to pass through a polycarbonate filter, which had been precoated with 2 mg/ml of Matrigel (Becton Dickinson, San Jose, CA). The lower chambers were filled with RPMI-1640 and 10% fetal bovine serum. Cells on the upper surface of the filters were wiped off after 48 h. The membranes were fixed using methanol for 15 min and stained using 0.5% crystal violet for 10 min. The cells on the underside of the filter were counted in five randomly selected microscopic views. All experiments were conducted three times in triplicate.
Colony formation assay
For the clonogenic assay, 5×102 cells were inoculated into 6-well plates and cultured for 14 d, with one medium change during this period. The samples were fixed for 30 min using 4% paraformaldehyde and stained using 0.5% crystal violet for 10 min. Photos were taken using a digital camera and colony counting was performed. All experiments were conducted three times in triplicate.
Subcutaneous tumor growth and lung colonization in vivo
SGC7901 cells (2×105 cells in 100 μl per mouse) stably transfected with shControl and shEIF3B-2 were injected into the left and right subperitoneal space of 6-week-old NOD/SCID mice (four mice in total), respectively. Tumor growth was monitored three times a week by measuring the length and width of the tumors using calipers.
To assess the effect of EIF3B on metastasis, SGC7901 cells stably transfected with shControl and shEIF3B-2 (1.5×106 cells in 100 μl per mouse) were injected intravenously via a 30-gauge needle inserted into the tail vein of 6-week-old female BALB/C-nude mice (eight mice in total, four mice per group). The mice were sacrificed 6 weeks later, and the lungs were removed and fixed using picric acid. The presence of lung metastases was evaluated at autopsy. All animal experiments involving in this manuscript were approved by the Committee of Laboratory Animal Ethics of Peking University of Oncology and the permit number is EAEC 2018–22. We also performed the standard guidelines of the Care and Use of Laboratory Animals of Peking University Beijing Cancer Hospital in all animal experiments.
Statistical analysis
All values were expressed as mean ± standard deviation. Chi-square tests were used to compare the differences in EIF3B protein expression. The overall survival and 5-year survival curve were calculated using the Kaplan–Meier method and analyzed using the Log-rank test. The relative risk (RR) of death associated with EIF3B expression and other predictor variables was estimated using the univariate Cox proportional hazards model. Multivariate Cox models were also constructed to estimate the RR for EIF3B expression. The Mann–Whitney test or Student’s T-test was used to compare cell function between two cell groups. All statistical analyses were conducted using the SPSS statistical package (version 20.0, SPSS Inc., Chicago, IL, USA). A two-tailed P-value less than 0.05 was considered to be statistically significant.
Results
The upregulation of EIF3B mRNA levels correlated with poor prognosis in GC
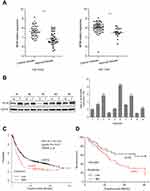
To investigate the role of EIF3B in GC, we first examined the expression of EIF3B in the Gene Expression Omnibus (GEO) database (GSE63089 and GSE13861), which indicated that EIF3B mRNA levels were significantly higher in GC tissues compared with matched adjacent normal mucosa, respectively (P<0.0001 and P<0.0001) (Figure 1A). We measured the expression levels of EIF3B protein and mRNA in five pairs and ten pairs of primary GC tissues and matched adjacent normal tissues, respectively. Consistent with the mRNA results, higher expression of EIF3B protein was detected in GC tissues compared with matched adjacent normal mucosa (Figure 1B). Kaplan–Meier survival analysis for EIF3B mRNA expression level and clinical outcomes in GC, using an online tool (http://kmplot.com/analysis), showed that the expression of EIF3B was significantly associated with poor overall survival in GC patients (Figure 1C). Kaplan–Meier survival analysis for EIF3B mRNA expression in 78 GC cases revealed the same relationship for 5-year survival (Figure 1D).
As shown in Table S1, although there was no significant correlation between EIF3B expression and age, gender, depth of invasion, lymph node metastasis, distant metastasis and TNM stage in GC patients, there was a tendency towards a correlation with TNM stage (P=0.072), which might be caused by the number of samples. High EIF3B expression predicted a shorter 5-year survival rate in GC patients, as assessed using the univariate Cox model (5-year survival rate: 62.5±0.96 vs 5.6±0.049, P=0.000, Table S2). Multivariate Cox regression analysis indicated that EIF3B expression was an independent poor prognostic factor for GC patients (RR =4.607 (2.208–9.611), P=0.000, Table S2).
Correlation of EIF3B protein expression with clinicopathological parameters and poor prognosis in GC patients
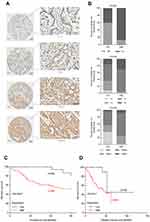
We analyzed the association between EIF3B protein expression and clinicopathological characteristics in 94 GC cases using IHC (Figure 2A). The distribution of staining intensity scores was as follows: 9 (9.6%) cases stained (-), 16 (17.0%) cases stained (+), 46 (48.9%) cases stained (++) and 23 (24.5%) cases stained (+++). High EIF3B protein expression was significantly associated with lymph node metastasis, depth of tumor invasion and TNM stage (P=0.000, 0.000 and 0.000, respectively) (Figure 2B and Table 1). Kaplan–Meier survival analysis for EIF3B protein expression indicated that patients with high EIF3B expression demonstrated significantly shorter 5-year survival and disease-free survival than those with low expression (P=0.0042 and 0.0017, respectively, by Log-rank test) (Figure 2C and D). The median overall survival time was 81.6±0.096 months for EIF3B low patients and 39.6±0.066 months for EIF3B high patients, respectively. High EIF3B protein expression predicted a shorter 5-year survival in GC patients, as assessed using the univariate Cox model (RR =4.810 (1.464–15.805), P=0.010, Table 2). Multivariate Cox regression analysis indicated that EIF3B expression was an independent poor prognostic factor for GC patients (RR =4.752 (1.352–16.706), P=0.015, Table 2).
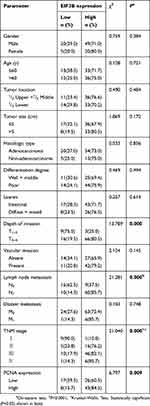 |
Table 1 Relationship between EIF3B expression and clinicopathological features in GC patients using IHC analysis |
 |
Table 2 Univariate and multivariate Cox proportional-hazards regression analysis for 5-year survival of GC patients using IHC analysis |
EIF3B promotes cell growth in vitro and in vivo
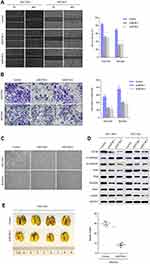
We measured the expression levels of EIF3B protein and mRNA in one gastric mucosa epithelial cell line (GES1), and six GC cell lines (N87, SGC7901, MKN28, AGS, BGC823 and MGC803), respectively. We found that EIF3B was highly expressed in five GC cell lines (N87, SGC7901, MKN28, BGC823 and MGC803), but low expressed in the AGS and GES1 cell lines (Figure 3A). To investigate the function of EIF3B in GC, SGC7901 and BGC823 cells were stably transfected with shControl, shEIF3B-1 and shEIF3B-2. Ectopic expression of EIF3B was confirmed through Western blot (Figure 3B). Down-regulation of EIF3B significantly inhibited cell proliferation and clonogenicity in SGC7901 and BGC823 cells (Figure 3C and D). Furthermore, high EIF3B protein expression was significantly associated with proliferating cell nuclear antigen (PCNA) expression (P=0.009, Table 1). We also found that EIF3B mRNA expression was significantly correlated with PCNA mRNA expression, as analyzed through gene expression profiling interactive analysis (GPEIA) (http://gepia.cancer-pku.cn/) (R =0.43, P=0.000) (Figure 3E).
To assess the tumorigenic ability of EIF3B, SGC7901 cells stably transfected with shControl and shEIF3B-2 were subcutaneously injected into the left and right subperitoneal space of NOD/SCID mice, respectively. Knockdown of EIF3B in SGC7901 cells markedly reduced the tumor size and weight of xenografts (Figure 3F). These in vitro and in vivo results support the tumor progression role of EIF3B in GC.
EIF3B promotes GC cell migration and invasion through epithelial-mesenchymal transition (EMT)
EIF3B knockdown in SGC7901 and BGC823 cells significantly prevented cell migration and invasion, as assessed through a wound healing assay and Matrigel invasion assay (both P<0.05; Figure 4A and B), respectively. Furthermore, EIF3B knockdown produced morphological changes in both SGC7901 and BGC823 cells, whereby the cells changed from a spindle shape to a plump shape, and protrusions were reduced or absent (Figure 4C). Knockdown of EIF3B in GC cells reduced the expression of mesenchymal related markers (N-cadherin, Snail, Slug, and Vimentin), promoted the expression of an epithelial marker (E-cadherin) and inactivated Stat3 signaling, as assessed through Western blot (Figure 4D).
Next, we evaluated the effect of EIF3B on tumor metastatic colonization in nude mice. SGC7901 cells stably transfected with shControl and shEIF3B-2 were injected into nude mice via the tail vein. After 6 weeks, the metastatic potential of the cells was assessed by counting colonized tumor nodules in the lungs of the mice. The EIF3B knockdown cell injected group had fewer lung tumor nodules compared with the control group (P<0.05, Figure 4E).
Upregulation of EIF3B enhanced the activity of PI3K/AKT/mTOR pathway signaling
Analyses of the EIF3B regulated gene signature via gene set enrichment analysis (GSEA) indicated that high expression of EIF3B was correlated with mTORC1 signaling pathway gene signatures (TCGA datasets and GSE21983; Figure 5A). These results suggest that EIF3B contributes to the activation of mTOR related pathway signaling. To validate these results, we confirmed through Western blot that PI3K, phospho-AKT and phosphor-mTOR are reduced in EIF3B knockdown cells (Figure 5B).
Discussion
GC progression involves multiple steps and is mediated by a series of genes, and translational control plays an important role.19 Several previous reports have indicated that elevated expression of EIF3B is highly correlated with oncogenesis in multiple tumors,20–22 but the role of EIF3B in GC was unclear and the molecular mechanisms had not been elucidated completely. In the current study, we first demonstrated that upregulation of EIF3B was significantly associated with an aggressive GC phenotype (lymph node metastasis, depth of tumor invasion and TNM stage) and with poor prognosis in GC patients. In addition, EIF3B significantly promoted GC cell growth, migration and invasion, and these results suggest that EIF3B is an oncogenic and prognostic biomarker for GC.
In general, EIF3B together with EIF3A could function as a nucleation core of the EIF3 complex, which is a key prerequisite in translation initiation, and downregulation of EIF3B led to a broad-spectrum low expression of all EIF3 subunits to 40–70% of the levels observed in control cells.23 The current study revealed that EIF3B possesses a strong tumor progression function in GC by promoting cell proliferation, migration and invasion in vitro. The growth-promotion effect of EIF3B was also demonstrated by its correlation with PCNA expression and xenograft tumor growth. A previous study suggested that overexpression of EIF3B could lead to malignant transformation of immortal fibroblasts, and its overexpression induced an oncogenic phenotype with an increased growth rate and protein synthetic rate, attenuated apoptosis and anchorage-independent growth.14
Knockdown of EIF3B using shRNA in GC cells significantly reduced their lung metastatic colonization in vivo. These results further confirmed the role of EIF3B in GC progression, and led us to explore the underlying biological mechanism of EIF3B in GC. Previous studies showed that downregulation of EIF3B can inhibit EMT to suppress cancer cell migration and invasion.15,17 The current study also indicated that knockdown of EIF3B led to alterations in cell morphology and the expression of EMT-associated proteins. Previous studies have reported that Stat3 plays a role in stimulating and controlling the transition of cells between epithelial and mesenchymal phenotypes,24 where it acts as a transcriptional activator of EMT-related genes in epithelial cancers.25 In the current study, we found that the phosphorylation of Stat3 was significantly decreased after EIF3B depletion, and the level of Snail and Slug, two targets of Stat3, were also downregulated accordingly. Together, our data suggest that EIF3B depletion could block the Stat3 pathway and inhibit EMT in GC.
The molecular mechanisms of EIF3B were predicted through an analysis of EIF3B regulated gene signatures via GSEA with TCGA and GEO21983 datasets. mTOR-related signaling may be closely related with EIF3B expression. We validated this pathway in vitro and confirmed an enhancement in PI3K, phospho-AKT and phospho-mTOR protein expression. mTOR is a serine/threonine protein kinase in the PI3K-related kinase family, that forms the catalytic subunit of two distinct protein complexes, known as mTOR complex 1 (mTORC1) and 2 (mTORC2).26 While mTORC1 regulates cell growth and metabolism, mTORC2 controls proliferation and survival.27 Previous studies have demonstrated that EIF3 upregulation combined with aberrant activation of the mTOR1 signaling pathway is common in cancer,28,29 and is essential for cancer development and metastasis.30 In the current study, we explored a regulatory mechanism of the PI3K/AKT/mTOR signaling axis mediated by EIF3B during GC progression. However, the function of AKT/mTOR during EIF3B upregulation requires further in vitro investigation using corresponding inhibitors. Interrupting the expression of translation initiation-related factors is a potential treatment strategy. As with EIF4 and its subunits,31 EIF3B may be a potential target for early therapeutic intervention to decrease the progression of GC.
Conclusion
In summary, the current results demonstrate that EIF3B is significantly upregulated in GC tissues and correlated with poor clinical outcomes. EIF3B demonstrated an oncogenic role in GC by promoting cell proliferation, migration and invasion in vitro, and stimulating xenograft tumor growth and lung metastasis in vivo. Moreover, EIF3B could regulate EMT process and the activity of the PI3K/AKT/mTOR signaling pathway in GC. These results suggest that EIF3B might be an early therapeutic target to inhibit the progression of GC and an independent prognostic biomarker for GC patients.
Acknowledgments
This study was supported in part by the National Natural Science Foundation of China (Grant Nos. 81502643 and 81802471), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (No. ZYLX201701), Beijing Cancer Hospital (support codes: A001538, 2017-23), the interdisciplinary medicine seed fund of Peking University (No. BMU2018MX018) and clinical medicine + X special fund from Peking University (support code: BMU2018XY010). The authors would like to thank Dr. Bin Dong and Dr. Zhongwu Li in the Department of Pathology at Peking University Cancer Hospital for excellent assistance in evaluating the IHC results.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. He W, Tu J, Huo Z, et al. Surgical interventions for gastric cancer: a review of systematic reviews. Int J Clin Exp Med. 2015;8(8):13657–13669.
3. Aoyama T, Yoshikawa T. Adjuvant therapy for locally advanced gastric cancer. Surg Today. 2017;47(11):1295–1302. doi:10.1007/s00595-017-1493-y
4. Sun KK, Shen XJ, Yang D, et al. MicroRNA-31 triggers G2/M cell cycle arrest, enhances the chemosensitivity and inhibits migration and invasion of human gastric cancer cells by downregulating the expression of zeste homolog 2 (ZH2). Arch Biochem Biophys. 2019;663:269–275. doi:10.1016/j.abb.2019.01.023
5. Duo-Ji MM, Ci-Ren BS, Long ZW, Zhang XH, Luo DL. Short-term efficacy of different chemotherapy regimens in the treatment of advanced gastric cancer: a network meta-analysis. Oncotarget. 2017;8(23):37896–37911. doi:10.18632/oncotarget.14664
6. Kanat O, O’Neil BH. Metastatic gastric cancer treatment: a little slow but worthy progress. Med Oncol. 2013;30(1):464. doi:10.1007/s12032-013-0464-4
7. Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40(2):228–237. doi:10.1016/j.molcel.2010.09.028
8. Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi:10.1016/j.cell.2009.01.042
9. Yu X, Zheng B, Chai R. Lentivirus-mediated knockdown of eukaryotic translation initiation factor 3 subunit D inhibits proliferation of HCT116 colon cancer cells. Biosci Rep. 2014;34(6):e00161. doi:10.1042/BSR20140078
10. Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi:10.1038/nrm2838
11. des Georges A, Dhote V, Kuhn L, et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525(7570):491–495. doi:10.1038/525S9a
12. Feng X, Li J, Liu P. The biological roles of translation initiation factor 3b. Int J Biol Sci. 2018;14(12):1630–1635. doi:10.7150/ijbs.24626
13. Liu Y, Neumann P, Kuhle B, et al. Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit. Structure. 2014;22(6):923–930. doi:10.1016/j.str.2014.03.010
14. Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282(8):5790–5800. doi:10.1074/jbc.M606284200
15. Zang Y, Zhang X, Yan L, et al. Eukaryotic translation initiation factor 3b is both a promising prognostic biomarker and a potential therapeutic target for patients with clear cell renal cell carcinoma. J Cancer. 2017;8(15):3049–3061. doi:10.7150/jca.19594
16. Xu F, Xu CZ, Gu J, et al. Eukaryotic translation initiation factor 3B accelerates the progression of esophageal squamous cell carcinoma by activating beta-catenin signaling pathway. Oncotarget. 2016;7(28):43401–43411.
17. Wang H, Ru Y, Sanchez-Carbayo M, Wang X, Kieft JS, Theodorescu D. Translation initiation factor eIF3b expression in human cancer and its role in tumor growth and lung colonization. Clin Cancer Res. 2013;19(11):2850–2860. doi:10.1158/1078-0432.CCR-12-3084
18. In H, Solsky I, Palis B, Langdon-Embry M, Ajani J, Sano T. Validation of the 8th Edition of the AJCC TNM staging system for gastric cancer using the national cancer database. Ann Surg Oncol. 2017;24(12):3683–3691. doi:10.1245/s10434-017-6078-x
19. Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–485. doi:10.1038/bjc.2012.494
20. Yin Y, Long J, Sun Y, et al. The function and clinical significance of eIF3 in cancer. Gene. 2018;673:130–133. doi:10.1016/j.gene.2018.06.034
21. Tian Y, Zhao K, Yuan L, et al. EIF3B correlates with advanced disease stages and poor prognosis, and it promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Cancer Biomark. 2018;23(2):291–300. doi:10.3233/CBM-181628
22. Golob-Schwarzl N, Krassnig S, Toeglhofer AM, et al. New liver cancer biomarkers: PI3K/AKT/mTOR pathway members and eukaryotic translation initiation factors. Eur J Cancer. 2017;83:56–70. doi:10.1016/j.ejca.2017.06.003
23. Wagner S, Herrmannova A, Sikrova D, Valasek LS. Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: the yeast-like core and the octamer. Nucleic Acids Res. 2016;44(22):10772–10788. doi:10.1093/nar/gkw972
24. Li B, Huang C. Regulation of EMT by STAT3 in gastrointestinal cancer (Review). Int J Oncol. 2017;50(3):753–767. doi:10.3892/ijo.2016.3762
25. Kang FB, Wang L, Jia HC, et al. B7-H3 promotes aggression and invasion of hepatocellular carcinoma by targeting epithelial-to-mesenchymal transition via JAK2/STAT3/Slug signaling pathway. Cancer Cell Int. 2015;15:45. doi:10.1186/s12935-015-0195-z
26. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi:10.1016/j.cell.2017.02.004
27. Xu K, Liu P, Wei W. mTOR signaling in tumorigenesis. Biochim Biophys Acta. 2014;1846(2):638–654.
28. Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–266. doi:10.1038/nrc2824
29. Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med (Berl). 2011;89(3):221–228. doi:10.1007/s00109-011-0726-6
30. Demosthenous C, Han JJ, Stenson MJ, et al. Translation initiation complex eIF4F is a therapeutic target for dual mTOR kinase inhibitors in non-Hodgkin lymphoma. Oncotarget. 2015;6(11):9488–9501. doi:10.18632/oncotarget.3378
31. Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi:10.1038/nrm3025
Supplementary materials
 |
Table S1 Relationship between RNA expression of EIF3B and clinicopathological features in GC patients using qPCR analysis |
 |
Table S2 Univariate and multivariate Cox proportional-hazards regression analysis for 5-year survival of GC patients using qPCR analysis |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.