Back to Journals » OncoTargets and Therapy » Volume 13
Eicosapentaenoic acid’s metabolism of 15-LOX-1 promotes the expression of miR-101 thus inhibits Cox2 pathway in colon cancer
Authors Cai Y, Liu J, Cai S, Miao E, Jia C, Fan Y, Li Y
Received 6 November 2019
Accepted for publication 6 April 2020
Published 15 June 2020 Volume 2020:13 Pages 5605—5616
DOI https://doi.org/10.2147/OTT.S237562
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Yi Cai,1,* Jie Liu,2,* Shao-kang Cai,1 Er-ya Miao,1 Cheng-qian Jia,1 Yong-zhi Fan,1 Ying-bo Li1
1Department of Pain Management, The Center Hospital of Wuhan, Tongji Medical College of Huazhong University of Science and Technology, Wuhan, Hubei, People’s Republic of China; 2Department of Pathology, The Center Hospital of Wuhan, Tongji Medical College of Huazhong University of Science and Technology, Wuhan, Hubei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ying-bo Li; Yong-zhi Fan
Department of Pain Management, The Center Hospital of Wuhan, Tongji Medical College of Huazhong University of Science and Technology, 26 Shengli Street, Jiang’an, Wuhan, Hubei 430014, People’s Republic of China
Email [email protected]; [email protected]
Purpose: It is well known that diet Eicosapentaenoic acid (EPA) is beneficial to colon cancer (CC). However, the underlying molecular mechanisms of EPA-relating miRNAs on genesis and development of this area is still unclear.
Materials and Methods: This study tries to find the function and specific role of EPA in CC through quantitative PCR (qPCR), Western blotting, immunofluorescence (IF), mass spectrometry, and immunohistochemistry (IHC) assays. By these methods, the enrichment of 15-LOX-1 metabolites of EPA, the expression of miR-101 and Cox2, and the relationship among them in CC are measured.
Results: The quantity of miR-101 was obviously suppressed in CC tissues and SW480 cells. After application of miR-101 mimics in CC cell lines, the Cox2 expression was inhibited too. Next, we confirmed that EPA could increase the expression of miR-101 induced by 15-LOX-1. Finally, we tested whether EPA functions as a regulator of miR-101 via the production of resolvin E3.
Conclusion: Our data demonstrate that the EPA– 15-LOX-1–miR-101-Cox2 signaling pathway owns a crucial position in the pathogenesis and development of diet-related CC. These findings exert exciting meanings for presenting new therapeutic angles in CC.
Keywords: colon cancer, 15-LOX-1, miR-101, Cox2, EPA
Introduction
Colon cancer (CC) is the third most common cancer and the cancer with highest mortality rate worldwide.1 Most CC cases are linked to diet, especially fatty acid-rich diets.2 It has been reported that eicosapentaenoic acid (EPA) exerts a rapid effect that may protect high-risk subjects from CC.3,4 Although the protective role of EPA against CC is gaining attention, the mechanism underlying this effect remains unclear. The function of all types of EPA metabolism has recently offered clues on the confirmation of EPA function.5,6
ALOX15 (also termed 15-LOX-1) is, like other lipoxygenases, an important enzyme for metabolizing polyunsaturated fatty acids to a wide range of physiologically and pathologically important products.7 Most studies on 15-LOX-1 focus on linoleic acid metabolites formed by 15-LOX-1 catalysis, such as 13-HODE,8 PGE2,9 and LXA4.10 Little is known about the role of resolvins in CC risk.11 A recent study found that resolvins could affect cholangiocarcinoma cell proliferation, but the specific mechanism still requires further exploration.12
In animals, resolvin E3, an EPA metabolite formed by ALOX15 catalysis, evokes anti-inflammatory and novel pro-resolving mechanisms as well as enhances microbial clearance,13 and alleviates hepatitis progression toward liver cancer.14 However, its role in CC has never been explored until now.
MicroRNAs (miRNAs) are a class of small noncoding RNAs regulating key cellular processes involved in cancer.15 Detection of miRNA patterns is important for the discovery of biomarkers for early cancer detection, diagnosis, and treatment.16–18 Some studies have indicated that fatty acid-mediated hsa-miR-32-5p inhibition is followed by BCL-2 and BCL2L11 mRNA suppression.19 Other studies have also shown that miRNA expression is associated with lipid, particularly fatty acid, metabolism.20,21
MiR-101, an important tumor-associated miRNA, provides novel insights into EP-4 receptor expression modulation at the post-transcriptional level, and therapeutic methods against miR-101 targets in CC.22 Therefore, the specific roles of miR-101 require further exploration to unravel complex pathogeneses and provide effective therapeutics for the patients with CC.
Here, we tested the miR-101 expression level and its relation to Cox2 in the CC cell line. We also discovered an association between EPA, resolvin E3, 15-LOX-1, and miR-101 in human CC cells. The role of resolvin E3 in CC and miR-101 expression requires further investigation.
Materials and Methods
C57BL/6J-ApcMin/J Mice Experiment
Mouse care and experimental protocols were approved by and conducted in accordance with the guidelines of the Animal Care and Use Committee of Wuhan central Hospital, China. C57BL/6J-ApcMin/J mice were purchased from the Jackson Lab (Cat# 002020, US). Each experimental group had 5 mice. At 4 weeks of age, the mice were fed with a 2% EPA diet or a control diet. Next, the mice were weighed each week. After 16 weeks on the diet, the mice were sacrificed, their colon cells were harvested, and the harvested cells were imaged.
Cell Culture and Cell Transfection
Human CC cell line SW480 was ordered from Typical Training Content Preservation Committee Cell Bank, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in McCoy 5ʹA medium (Thermo Fisher Scientific, MA, USA) with 10% FBS (Thermo Fisher Scientific, MA, USA) and penicillin/streptomycin (100 U/mL) at 37°C. In addition, mimics and inhibitors targeting miR-101 and negative controls (NCs) were acquired from Ribo Bio Co., Ltd. Cells were transfected with RNAs or NCs using Lipofectamine 3000 (Invitrogen, CA, USA) according to the manufacturer’s protocol.
EPA-free fatty acid (EPA-FFA, ALFA, SLA Pharma AG, Switzerland; 150 µM) or 100% ethanol as a control was applied to SW480 cells for 48 hours in vitro assay.
RNA Isolation and Real-Time PCR (qRT-PCR) Assays
Total RNA was extracted from cell lines using Trizol reagent (Invitrogen, CA, USA). The indicated miRNAs were reverse transcribed using Prime Script™ RT-PCR Kit (Takara, Tokyo, Japan) and used as template in a qRT-PCR with Real-time PCR Master Mix (TOYOBO, Japan) and measured by Step One Real-Time PCR System (Applied Biosystems, CA, USA). U6 RNA was used as the endogenous control. PCR was performed with primers listed in Table S1. All experiments were performed in triplicate.
CCK-8 Assay
Transfected or control SW480 cells (2000 per well) were seeded in a 96-well plate and incubated at 37°C for 24 hours. Cell viability was analyzed at 24 h, 48 h, and 72 h using the Cell Counting Kit-8 (CCK-8; Kumamoto, Japan) according to the manufacturer’s protocols.
Western Blotting Assay
All proteins were extracted from CC tissues and cells, and protein concentrations were measured using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, MA, USA). Specific primary antibodies were used against Cox2 (# 4842, 1:1000; CST, USA) and β-actin (# sc-47778, 1:1000; Santa Cruz, CA, USA), and HRP secondary antibodies (# 31430 and # 31460, 1:4000; Thermo Fisher Scientific, MA, USA) were used for signal amplification. The protein bands were developed using the chemiluminescence detection system (Thermo Fisher Scientific, MA, USA). All Western blot data presented are representative of three independent experiments.
Soft Agar Colony Formation Assay
Soft agar colony formation assay was performed using transfected and control cells. Briefly, 1 × 103 cells were equally seeded in a six-well plate containing 0.3% noble agar and were grown for 7 days. The number of colonies was determined by direct counting under an inverted microscope.
Plasmid Construction
The full-length 3ʹ-UTR of Cox2 was amplified using PCR and cloned into the HindIII and SpeI cloning sites of the pMIR-REPORT luciferase expression vector (Ambion, USA). The predicted miR-101 binding sequences in the Cox2 3ʹ-UTR were mutated in the Luc-MUT vector using site-directed mutagenesis (Stratagene, USA).
The 15LOX-1 plasmid was purchased from Origene (#: RG206621). An empty pCMV6 vector was used as the control.
Patients and Specimens
Fifty human CC tissues and corresponding nearby non-tumor colon tissues (5-cm away from the tumor margin) were harvested from patients who underwent colon resection between January 2014 and November 2016 at the General Surgery department of Wuhan central Hospital affiliated to Huazhong University of Science and Technology. Written informed consent was obtained from all patients and healthy volunteers before beginning the study. The study was approved by the medical ethics committee of Wuhan central Hospital, China.
Luciferase Reporter Assay
To confirm the miR-101 target sites, sW480 cells were co-transfected with miR-101 and luciferase reporter constructs with wild-type or mutated Cox2 3ʹ-UTR. The signals were tested 24 hours after transfection using the Dual-Luciferase Assay Kit (Promega, WI, USA).
Liquid Chromatography–Mass Spectrum (LC/MS) Assays
15-LOX-1-transfected SW480 and control cell lysates were used as previously described in LC/MS according to the manufacturer’s instructions to measure the resolvin E3 enrichment after EPA application.23
Statistical Analysis
Statistical significance was determined using the unpaired Student’s t-test, chi-square test, or analysis of variance (one-way or two-way) with Bonferroni adjustments for multiple comparisons. All tests were two-sided, and significance was defined as P<0.05. All acquired data were analyzed using the SPSS statistical software (version 16.0).
Results
miR-101 Expression Is Suppressed in Colon Cancer Tissues Compared to That in Nearby Normal Tissues and Negatively Associated with Cox2 Expression
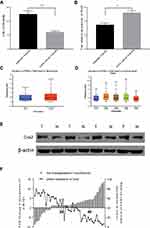
miR-101 expression was measured using Quantitative Real-time PCR in 50 tumor and paired adjacent colon tissues from the patients with CC. miR-101 expression was significantly downregulated in CC tissues compared to that in normal colon tissues (P<0.0001; Figure 1A).
We measured Cox2 expression in the same tissues using qPCR and Western blotting. We showed that the difference in Cox2 mRNA levels between the tissues was not as obvious as the difference in miR-101 expression (Figure 1B). Based on the TGCA database, COX2 (PTGS2) expression was much higher in CC tissues than that in normal tissues and was the highest in the early stage (Stage 1) (Figure 1C and D). Furthermore, the Cox2 protein level in cancer tissues was higher than that in control tissues (Figure 1E). Cox2 expression was strongly negatively associated with miR-101 expression (Figure 1F).
miR-101 Expression Manipulation Affects SW480 Cell Viability
To explore the effect of miR-101 overexpression on CC cells, miR-101 mimics and control siRNA were used in subsequent experiments. qRT-PCR results verified that miR-101-overexpressing SW480 cells had a much higher miR-101 level than the control cells 48 h post-transfection (P<0.01, Figure 2A). The CCK-8 assay revealed that miR-101 upregulation inhibited SW480 cell viability (Figure 2B).
Conversely, miR-101 inhibitors and control siRNA were used to observe their effects on SW480 cells. qRT-PCR results confirmed that miR-101 inhibition led to decreased miR-101 levels compared to those in control cells 48 h post-transfection (P<0.01, Figure 2C). The CCK-8 assay revealed that miR-101 downregulation promoted SW480 cell viability (Figure 2D).
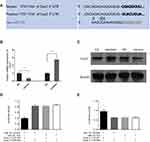
miR-101 Directly Regulates Cox2 in SW480 Cells
To confirm the miR-101 target in CC, we searched the miRNA target prediction databases (MiRDB and TargetScan) to find available target genes. Cox2 was predicted to be a target by both databases. The putative sequence of has-miR-101 binding site in the 3′ -UTR of Cox2 is shown in Figure 3A. To confirm whether Cox2 was affected by miR-101, we used Western blotting and qRT-PCR in miR-101 overexpressed or silenced and control cells. Only Cox2 protein expression levels were downregulated or upregulated in response to miR-101 expression manipulation (Figure 3B and C). To further test whether Cox2 is a direct miR-101 target, we constructed a wild-type Cox2 3′-UTR luciferase reporter vector. miR-101 overexpression suppressed luciferase activity in the wild-type Cox2 3′-UTR luciferase reporter construct, which was negated when the verified miR-101 binding sequences in the Cox2 3′-UTR were mutated, showing that miR-101 inhibits Cox2 via the predicted sequence (Figure 3D). All of the above results verified that miR-101 directly affects Cox2 expression in SW480 cells in vitro.
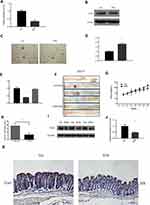
EPA Promotes miR-101 Activation and Downregulates Cox2 Both in vitro and in vivo
We used qPCR and Western blotting to analyze whether EPA application could decrease Cox2 expression in SW480 cells (Figure 4A and B). The soft agar clone formation assay further confirmed the effect of EPA on inhibiting the proliferation of SW480 cells (Figure 4C). Cox2 inhibition was accompanied by miR-101 upregulation (Figure 4D). The effect of EPA on Cox2 was negated when miR-101 expression was inhibited by an inhibitor (Figure 4E). The results show that EPA manipulates miR-101 expression to affect Cox2 levels. We further confirmed this result in Apc mutant mice. Four-week-old Apc mutant mice were fed 1% EPA diet for 16 weeks and the tumor size and number were found to be decreased in the EPA-fed mice compared to the control mice (Figure 4F, G, and H). Cox2 expression was also similar in SW480 and mouse colon cells (Figure 4I–K).
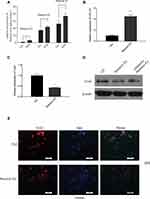
Resolvin E3 Inactivates the Cox2 Signaling Pathway by Increasing miR-101 Expression
Resolvin Es is a group of specialized pro-resolving mediators, which can evoke anti-inflammatory and novel pro-resolving mechanisms, enhance microbial clearance,24 and alleviate hepatitis progression toward liver cancer14 in animals. We observed the levels of EPA metabolites associated with cancer inhibition (Resolvin Es) in SW480 cells compared to those in control cells using Liquid chromatography-tandem mass spectrometry (LC/MS) to identify the factors involved in EPA function on miR-101 expression. Resolvin E3 was the most notable lipid metabolite with different levels between the two groups (Figure 5A). Additionally, we found that it increased miR-101 expression (Figure 5B) and inhibited Cox2 expression (Figure 5C and E). Resolvin E3 activity was strongly inhibited when miR-101 expression was inhibited (Figure 5D).
Our data suggested that resolvin E3 promotes miR-101-induced Cox2 pathway inactivation.
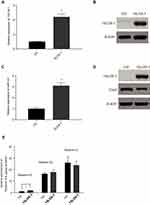
15-LOX-1 Enhances the Effect of EPA on Cox2 and miR-101 Expression
Since resolvin E3 is the main metabolite formed by 15-LOX-1 catalysis, we tested the role of this gene in SW480 cells to confirm its effect. We transduced the 15-LOX-1 overexpression plasmid and control vector into SW480 cells and confirmed the transfection effect using qPCR and Western blotting (Figure 6A and B). We used qPCR to show that miR-101 expression was increased (Figure 6C). Western blotting results also supported our conclusion that Cox2 expression was downregulated by 15-LOX-1 expression (Figure 6D). We observed the levels of EPA metabolites between control and 15-LOX-1 over-expressed SW480 cells using LC/MS to further confirm the relationship between Resolvin Es and 15-LOX-1. Resolvin E3 was the most notable lipid metabolite with different levels between the two groups (Figure 6E).
Discussion
miRNAs are pivotal mediators of carcinogenesis and CC progression.25,26 To our knowledge, the mechanism underlying dietary induction of CC is still unclear. In a previous study,27 we confirmed the strong association between miRNA expression and CC carcinogenesis. Among all the identified miRNAs, we focused on miR-101 since it was reportedly related to Cox2 in CC.22 Until now, we wondered about the factor affecting miRNA-101 expression to affect Cox2 function. Our data showed that miR-101 expression was strikingly downregulated in CC tissues compared to that in nearby normal tissues. It was also remarkably associated with EPA application, which plays a role in producing resolvin E3, the EPA metabolite formed by 15-LOX-1 catalysis. Thus, the marked association between miR-101 and dietary EPA may provide a new clue to cure CC in the early stages.
Fatty acids are very important constituents in our daily diet and involved in different diseases.28,29 Increasing evidence depicts an important role of polyunsaturated fatty acids (PUFAs), especially EPA, in rearranging the expression of many genes that could contribute to CC carcinogenesis.30 Lee et al reported the chemopreventive and chemotherapeutic effects of EPA on colon carcinogenesis.31 Kansal et al analyzed the COX-2 expression levels, alterations in Wnt signaling, and PPARγ expression to clarify the role of lipid-mediated signaling events by EPA in a colon cancer model. However, a clear relationship between EPA and miR-101 is shown in our study. We demonstrated that Cox2 is the direct target gene of miR-101 by manipulating miR-101 expression. We found that EPA could enhance the effect of miR-101 on Cox2. This is the first time that EPA was linked to Cox2 via miRNAs. Further, we found that resolvin E3, an EPA metabolite formed by 15LOX-1 catalysis, affects miR-101 expression to suppress Cox2 function. Resolvin E3 has been confirmed to inhibit tumors in several cancers, but its effects on CC have not been studied yet. These results were verified in vivo. After feeding Apc mutant mice with EPA diet for 14 weeks, we found that the tumor number and volume in the EPA-fed mice decreased compared to that in the control mice. When we detected the lipid profiles in colon cells of this batch of mice, the quantity of resolvin E3 was most different among them. The Cox2 and miR-101 protein levels were measured. All results verified those of our previous study. As expected, our data showed that resolvin E3 inhibits Cox2 expression via an miR-101-mediated pathway.
15-LOX-1 is an important factor in fatty acid metabolism,32 and a previous study has mostly focused on 13-HODE.33 The role of 15-LOX-1 as a catalyst metabolizing the dietary fatty acid EPA has been neglected for a long time. In particular, resolvin E3, the main EPA metabolite formed by 15-LOX-1 catalysis, has not been extensively studied. Here, we first determined the relationship between resolvin E3 and miRNAs-miR-101. We further confirmed our hypothesis using 15-LOX-1 overexpression. Resolvin E3 level decreased when 15-LOX-1 was overexpressed and miR-101 and Cox2 expression also changed at the protein and mRNA levels. Our data emphasize the critical role of EPA in CC carcinogenesis and progression. We found that resolvin E3, an important EPA metabolite, could regulate miRNA expression and manipulate Cox2 expression.
To our knowledge, abnormal miR-101 expression in CC was reported several years ago. However, the EPA–15-LOX-1–Resolvin E3–miR-101–Cox2 signaling pathway regulation axis has not been reported before. This may help the researchers understand the mechanisms of diet-related CC. Further studies are needed to investigate the mechanism of the effect of resolvin E3 on miR-101 expression to elucidate the relationship between lipids and complex miRNA regulation network in CC.
Disclosure
The authors report no funding and no conflicts of interest for this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Meyerhardt JA, Niedzwiecki D, Hollis D, et al. Association of dietary patterns with cancer recurrence and survival in patients with stage III colon cancer. JAMA. 2007;298(7):754–764. doi:10.1001/jama.298.7.754
3. Anti M, Marra G, Armelao F, et al. Effect of omega-3 fatty acids on rectal mucosal cell proliferation in subjects at risk for colon cancer. Gastroenterology. 1992;103(3):883–891. doi:10.1016/0016-5085(92)90021-P
4. Fuentes NR, Mlih M, Barhoumi R, et al. Long-Chain n-3 fatty acids attenuate oncogenic KRas-driven proliferation by altering plasma membrane nanoscale proteolipid composition. Cancer Res. 2018;78(14):3899–3912. doi:10.1158/0008-5472.CAN-18-0324
5. Lee CR, Zeldin DC. Resolvin infectious inflammation by targeting the host response. N Engl J Med. 2015;373(22):2183–2185. doi:10.1056/NEJMcibr1511280
6. Ash C. Protectin and resolvin gut inflammation. Science. 2017;356(6334):150–151. doi:10.1126/science.356.6334.150-d
7. Kristjansson RP, Benonisdottir S, Davidsson OB, et al. A loss-of-function variant in ALOX15 protects against nasal polyps and chronic rhinosinusitis. Nat Genet. 2019;51(2):267–276. doi:10.1038/s41588-018-0314-6
8. Zuo X, Peng Z, Wu Y, et al. Effects of gut-targeted 15-LOX-1 transgene expression on colonic tumorigenesis in mice. J Natl Cancer Inst. 2012;104(9):709–716. doi:10.1093/jnci/djs187
9. Schroeder CP, Yang P, Newman RA, Lotan R. Eicosanoid metabolism in squamous cell carcinoma cell lines derived from primary and metastatic head and neck cancer and its modulation by celecoxib. Cancer Biol Ther. 2004;3(9):847–852. doi:10.4161/cbt.3.9.1037
10. Green AR, Freedman C, Tena J, et al. 5 S,15 S-Dihydroperoxyeicosatetraenoic acid (5,15-diHpETE) as a Lipoxin Intermediate: reactivity and kinetics with human leukocyte 5-lipoxygenase, platelet 12-lipoxygenase, and reticulocyte 15-lipoxygenase-1. Biochemistry. 2018;57:6726–6734. doi:10.1021/acs.biochem.8b00889
11. Werz O, Gerstmeier J, Libreros S, et al. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat Commun. 2018;9(1):59. doi:10.1038/s41467-017-02538-5
12. Khophai S, Thanee M, Techasen A, et al. Zileuton suppresses cholangiocarcinoma cell proliferation and migration through inhibition of the Akt signaling pathway. Onco Targets Ther. 2018;11:7019–7029. doi:10.2147/OTT.S178942
13. Isobe Y, Arita M, Iwamoto R, et al. Stereochemical assignment and anti-inflammatory properties of the omega-3 lipid mediator resolvin E3. J Biochem. 2013;153(4):355–360. doi:10.1093/jb/mvs151
14. Kuang H, Hua X, Zhou J, Yang R. Resolvin D1 and E1 alleviate the progress of hepatitis toward liver cancer in long-term concanavalin A-induced mice through inhibition of NF-kappaB activity. Oncol Rep. 2016;35(1):307–317. doi:10.3892/or.2015.4389
15. Callegari E, Gramantieri L, Domenicali M, D’Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22(1):46–57. doi:10.1038/cdd.2014.136
16. Zhou SJ, Deng YL, Liang HF, Jaoude JC, Liu FY. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ. 2017;24(9):1577–1587. doi:10.1038/cdd.2017.87
17. Liu FY, Zhou SJ, Deng YL, et al. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis. 2015;6:e1670. doi:10.1038/cddis.2015.46
18. Zhou SJ, Liu FY, Zhang AH, et al. MicroRNA-199b-5p attenuates TGF-beta1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Br J Cancer. 2017;117(2):233–244. doi:10.1038/bjc.2017.164
19. Kopke S, Buhrke T, Lampen A. miRNA expression in human intestinal Caco-2 cells is comparably regulated by cis- and trans-fatty acids. Lipids. 2015;50(3):227–239. doi:10.1007/s11745-015-3988-x
20. Davidson LA, Wang N, Shah MS, Lupton JR, Ivanov I, Chapkin RS. n-3 Polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis. 2009;30(12):2077–2084. doi:10.1093/carcin/bgp245
21. Malcomson FC, Willis ND, Mathers JC. Is resistant starch protective against colorectal cancer via modulation of the WNT signalling pathway? Proc Nutr Soc. 2015;74(3):282–291. doi:10.1017/S002966511500004X
22. Chandramouli A, Onyeagucha BC, Mercado-Pimentel ME, et al. MicroRNA-101 (miR-101) post-transcriptionally regulates the expression of EP4 receptor in colon cancers. Cancer Biol Ther. 2012;13(3):175–183. doi:10.4161/cbt.13.3.18874
23. Barden AE, Shinde S, Burke V, et al. The effect of n-3 fatty acids and coenzyme Q10 supplementation on neutrophil leukotrienes, mediators of inflammation resolution and myeloperoxidase in chronic kidney disease. Prostaglandins Other Lipid Mediat. 2018;136:1–8. doi:10.1016/j.prostaglandins.2018.03.002
24. Serhan CN, Chiang N, Dalli J. The resolution code of acute inflammation: novel pro-resolving lipid mediators in resolution. Semin Immunol. 2015;27(3):200–215. doi:10.1016/j.smim.2015.03.004
25. Chivukula RR, Shi G, Acharya A, et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 2014;157(5):1104–1116. doi:10.1016/j.cell.2014.03.055
26. Valeri N, Braconi C, Gasparini P, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469–483. doi:10.1016/j.ccr.2014.03.006
27. Hutchison J, Cohen Z, Onyeagucha BC, Funk J, Nelson MA. How microRNAs influence both hereditary and inflammatory-mediated colon cancers. Cancer Genet. 2013;206(9–10):309–316. doi:10.1016/j.cancergen.2013.06.005
28. Sergeant S, Ruczinski I, Ivester P, et al. Impact of methods used to express levels of circulating fatty acids on the degree and direction of associations with blood lipids in humans. Br J Nutr. 2016;115(2):251–261. doi:10.1017/S0007114515004341
29. Silva Figueiredo P, Carla Inada A, Marcelino G, et al. Fatty acids consumption: the role metabolic aspects involved in obesity and its associated disorders. Nutrients. 2017;9(10). doi:10.3390/nu9101158.
30. Colby JK, Jaoude J, Liu F, Shureiqi I. Oxygenated lipid signaling in tumor-associated macrophages-focus on colon cancer. Cancer Metastasis Rev. 2018.
31. Lee JY, Sim TB, Lee JE, Na HK. Chemopreventive and chemotherapeutic effects of fish oil derived omega-3 polyunsaturated fatty acids on colon carcinogenesis. Clin Nutr Res. 2017;6(3):147–160. doi:10.7762/cnr.2017.6.3.147
32. D’Eliseo D, Velotti F. Omega-3 fatty acids and cancer cell cytotoxicity: implications for multi-targeted cancer therapy. J Clin Med. 2016;5:2. doi:10.3390/jcm5020015
33. Wendell SG, Golin-Bisello F, Wenzel S, Sobol RW, Holguin F, Freeman BA. 15-Hydroxyprostaglandin dehydrogenase generation of electrophilic lipid signaling mediators from hydroxy omega-3 fatty acids. J Biol Chem. 2015;290(9):5868–5880. doi:10.1074/jbc.M114.635151
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.