Back to Journals » OncoTargets and Therapy » Volume 10
EGFR with TKI-sensitive mutations in exon 19 is highly expressed and frequently detected in Chinese patients with lung squamous carcinoma
Authors Memon AA, Zhang HP, Gu Y, Luo Q, Shi JJ, Deng Z, Ma J, Ma W
Received 16 December 2016
Accepted for publication 17 April 2017
Published 18 September 2017 Volume 2017:10 Pages 4607—4613
DOI https://doi.org/10.2147/OTT.S130051
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Aadil Ahmed Memon,1 Haiping Zhang,2 Ye Gu,3 Qian Luo,4 Jiajun Shi,1 Zixin Deng,1 Jian Ma,5 Wei Ma1
1State Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, 2Oncology Department, 3Endoscope Department, Shanghai Pulmonary Hospital, Tongji University School of Medicine, 4Core Facility and Technical Service Center, School of Life Science and Biotechnology, Shanghai Jiao Tong University, 5Pneumology Department, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China
Abstract: Recently, tyrosine kinase inhibitors (TKIs) have been recommended as a first-line treatment for advanced non-small cell lung cancer (NSCLC), significantly improving the treatment outcomes of lung adenocarcinoma patients with the EGFR mutation. However, the application of TKIs for lung squamous cell carcinoma (SCC), the second largest pathological subtype of NSCLC, remains controversial because available data for the EGFR mutation profile and frequency in SCC patients are limited. In this study, 89 bronchoscopic–biopsy specimens from Chinese SCC male patients were assayed for EGFR exon 19 mutation, using improved polymerase chain reaction-denature gel gradient electrophoresis. EGFR exon 19 mutations were detected in 77 of 89 (86.5%) patients, and included six kinds of point mutations (11.6%) and two deletions (Del_747-751 [64.9%] and Del_746-751 [23.3%]). We found that the proportion of mutated EGFR varied from 0.98% to 100% in positive specimens and increased with the development of the disease. The difference of proportion between Stage IV patients and Stage II patients or Stage III patients was significant (P<0.001). These results provided valuable clues to explain the reason why patients harboring the same mutation responded distinctly to TKI treatment. Del_747-751 and Del_746-751 were the dominant mutations in the assayed SCC patients (76.4%), and both belong to the EGFR–TKI-sensitive mutation. Recently research demonstrated that Del_746-751 patients have better response to EGFR-TKI than Del_L747-751 patients. However, our study indicated that majority of SCC patients (55.5%) carried Del_ L747-751. We suggest that the unique clinic features of SCC should be further studied to reveal the mechanism of poorer treatment outcome of EGFR–TKI therapy, and that a better treatment plan and more specific, potent targeted drugs for lung SCC need to be developed.
Keywords: lung squamous carcinoma, EGFR exon 19, mutation profile, PCR-DGGE, tyrosine kinase inhibitors
Introduction
Lung cancer is a malignancy associated with high mortality, and the situation is especially serious in China compared to the world. In 2015, 733,300 new lung cancer cases were reported and 610,200 lung cancer-related deaths occurred in China.1 The World Health Organization (WHO) predicts that annual number of deaths from lung cancer in China is estimated to reach 1 million by 2025.2 Approximately 70%–80% of lung cancer is non-small cell lung cancer (NSCLC), and squamous cell carcinoma (SCC) is the second largest subgroup of NSCLC.3–5 Tyrosine kinase inhibitors (TKIs) have been recommended as a first-line treatment option for advanced adenocarcinoma patients, and have significantly improved the treatment outcomes for patients.6 EGFR–TKIs combined with chemotherapy were approved as a second-line treatment option in USA and Europe for advanced squamous carcinoma patients.6 Currently, SCC patients still have limited treatment options in addition to chemotherapy,7 thus resulting in a poorer treatment outcome as compared to adenocarcinoma patients.7,8
The EGFR signaling pathway is associated with several key cellular metabolic processes.9–12 An aberrant EGFR pathway contributes crucially to tumor initiation and progress.12 EGFR mutations can be identified in 80% of NSCLC patients,13 especially in the Asian race, never smokers, women, and patients with adenocarcinoma,10 therefore adenocarcinoma patients with EGFR mutation can benefit from TKI therapy.12 The mutation frequency of EGFR in SCC was reported to be only ~1%–15%,14 and EGFR mutation analysis and TKI treatment are not routinely recommended for SCC patients.14,15 For example, the European Society for Medical Oncology (ESMO) recommended the EGFR mutation test for nonsquamous carcinoma patients.16 However, these recommendations were based on limited data on the EGFR mutation profile and frequency of SCC, especially in Chinese and other Asian populations.7,15 When we searched the PubMed database using keywords “EGFR” and “lung squamous carcinoma,” few results were returned.
Recently, several studies demonstrated that the SCC patients could benefit from TKI therapy,17,18 although the treatment outcome, the progression-free survival (PFS), and overall survival (OS) were not as good as those of the adenocarcinoma patients.18 Furthermore, the discrepancy among different studies and/or patients was significant. For example, Xu et al19 reported 26 Chinese SCC EGFR mutation-positive patients who were treated with TKI, and the PFS was 3.98 months (95% CI, 3.22–4.63 months). In another study,20 53 Japanese SCC patients were treated with TKI, and the median PFS of EGFR-positive patients (n=20) was 1.4 months (95% CI, 0.7–5.8 months), while the PFS of EGFR-negative patients (n=33) was 1.8 months (95% CI, 1.0–2.4 months). Though no statistical significance in median PSF was detected between EGFR-positive and −negative groups (P=0.1734), the range of PFS of EGFR-positive (8.3-fold) patients is much wider than that of EGFR-negative patients (2.4-fold),20 which suggested that greater interindividual difference existed in EGFR-positive patients.
The EGFR mutational profile was obtained by direct sequencing,21 high-throughput sequencing,22,23 and some polymerase chain reaction (PCR)-based methods,20 which will ultimately determine whether or not the TKI12 treatment can be applied to specific patients. However, the abovementioned methods have some ramifications in terms of sensitivity, cost, and time24 and, as a result, no method is highly recommended for regular clinical diagnosis of lung cancer.25–28 Most studies detected the EGFR mutation using genomic DNA extracted from paraffin specimens.24 EGFR was overexpressed in advanced-stage NSCLC patients at both protein and mRNA levels,29 but the ratio of mutated EGFR to wild type expressed in SCC still needs to be determined.
This study aimed to investigate the mutation status of EGFR exon 19 by PCR-denature gel gradient electrophoresis (DGGE) to provide information for understanding the mechanism of poorer treatment outcome of EGFR–TKI therapy for SCC patients by providing a clear view of the mutation profile and frequency in Chinese SCC patients.
Materials and methods
In this study, we assayed EGFR mutations in Chinese patients with lung squamous carcinoma using PCR-DGGE and direct sequencing of PCR products.
Sample collection
Specimens were collected from patients who underwent bronchoscopic biopsy for lung cancer in Shanghai Pulmonary Hospital from 2013 to 2015, with the approval of the Ethics committee of Tongji University. All patients involved in this study provided written informed consent for the use of their tissue samples in this study. The specimens that were confirmed to be SCC by pathological examination were used for the following experiments. Detailed clinical information is shown in Table 1. All specimens were immediately preserved in the RNA store reagent (Tiangen Biotech, Beijing, China) at 4°C until total RNA extraction.
  | Table 1 Clinicopathological features of patients with lung squamous cell carcinoma |
Chemical and reagents
The chemicals used in this study were purchased from Sigma-Aldrich (St Louis, MO, USA).
Total RNA extraction and reverse transcription
Total RNA Extraction Kit (Tiangen Biotech) was used to extract the total RNA. Approximately 1,000 ng of total RNA was reverse transcribed in 20 μL volume using FastQuant cDNA first chain synthesis Kit (Tiangen Biotech) and the manufacturer’s instructions were followed throughout the operation. cDNA were stored at −20°C for future experiments.
PCR and DGGE
PCR and DGGE were performed according to the methods reported by Shahi et al,24 with two modifications, ie, 20 cycles for first-round PCR, and use of cDNA as the template for PCR instead of gDNA. Universe High-Fidelity Hot Start DNA Polymerase (Biotool, Houston, TX, USA) was used as the PCR enzyme. DGGE analysis was performed on the Universal Mutation detection system (D-Code™; Bio-Rad, Hercules, CA, USA). A 30%–60% denaturation gradient gel was used, and samples were allowed to run for 16 hours at 120 V at 58°C. The gels were stained with GelRed (GelRed™; Biotium, Fremont, CA, USA) and images were documented using Gel Documentary XR+ System (Gel Doc™ XR; Bio-Rad) and quantified using Gel-pro Analyzer 4.0 (Media Cybernetics, Rockville, MD, USA). The samples were analyzed in triplicate and always with positive and negative controls (Figure 1).
Sequence analysis
After DGGE, each candidate band was cut from the gel, crushed, and soaked in 30 μL Tris–EDTA buffer overnight at 4°C, individually, and then the buffer was used as a template for cloning PCR. PCR primers were same as PCR-DGGE without the GC clamp. PCR products were cloned using Lethal Based Fast Cloning Kit (Tiangen Biotech) and sequenced by Sunny Biotechnology Co. (Shanghai, China). Sequencing results were analyzed by BLAST.
Statistical analysis
Statistical analysis was performed with SPSS 17.0 or GraphPad Prism 5.0. Two groups of data were analyzed using unpaired t-test to determine the significance. For three or more groups of data, one-way analysis of variance was adopted. Results were presented as mean ± SD. P<0.05 was considered statistically significant.
Results
Characteristics of patients involved in this study
Since no sufficient specimens from female SCC patients have accumulated due to the low incidence of SCC in the Chinese female population, only the specimens from male patients were analyzed in this study. The detailed characteristics such as patients’ age, smoking history, and tumor node metastasis stage are shown in Table 1.
Mutation profile of EGFR exon 19
In 89 SCC specimens assayed, 41 patients (46%) were in early stage (I–II) and 48 were in advanced stage (III–IV) (54%). Two kinds of deletions (c.2240-2254 and c.2237-2251) and six point mutations were identified in EGFR exon 19 regions. After being verified by the database of catalogue of somatic mutations in cancer (COSMIC) (http://cancer.sanger.ac.uk/cosmic) (Table 2), five point mutations were identified for the first time in all kinds of cancer (Table 2). In early-stage patients (I–II), seven different kinds of mutations were identified, whereas only four kinds of mutations were found in advanced-stage patients (III–IV) (Tables 2 and 3). Fifty of 89 patients (56.2%) harbored the c.2240-2254 deletion (Table 2) and 18 patients (20.2%) harbored the c.2237-2251 deletion (Table 2), which accounted for 76.4% of the total number of SCC patients studied; and both deletions are EGFR–TKI-sensitive.30 Nine out of 89 patients (11.6%) were found to have point mutations; three of these patients were identified to have either a double mutation or a combination of point mutations. Twelve patients (15.5%) carried no mutation in exon 19 (Table 3).
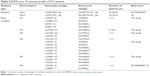  | Table 2 EGFR exon 19 mutation profile of SCC patients |
  | Table 3 Distribution of eight kinds of mutation in different SCC stages |
In Stage I, 50% of patients harbored deletion 1 (n=4). In Stage II patients, 48.6% of patients had deletion 1 (n=18), and 24.3% had deletion 2 (n=9). For Stage III patients, 68.1% of patients (n=15) carried deletion 1 and 4.5% of patients (n=1) had deletion 2. For Stage IV patients, 57.6% (n=15) of patients harbored deletion 1 and 30.7% carried deletion 2 (n=8) (Table 3).
Point mutations in EGFR exon 19 were identified in 25% of Stage I patients (n=4), 13.5% of Stage II patients (n=37), and 9.1% of Stage III patients (n=22). Two kinds of point mutations were identified in three patients, and no point mutation was detected in any of the Stage IV patients assayed (n=26) (Table 3; Figure 1B).
Among the 89 patients involved in this study, there were 45 smokers, 35 non-smokers, and the remaining 9 patients did not have smoking information. For smoking patients, their average smoking history was 34.4±10.7 years and the average daily consumption was 30.7±13.8 cigarettes/day (Table 4). Compared with smoking patients (42 patients [93.3%]), nonsmoking patients had a much lower incidence of the exon 19 mutation (27 patients [77.1%]). Furthermore, 7 (15.5%) smoking patients and only 2 (5.7%) nonsmoking patients were identified as harboring point mutations. But no significant difference was observed in mutation frequency and mutation type between smoking and nonsmoking patients (Table 4).
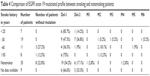  | Table 4 Comparison of EGFR exon 19 mutational profile between smoking and nonsmoking patients |
Quantification of DGGE bands
Each patient’s wild type and mutated EGFR band in DGGE gel was quantified, and the proportion of mutated EGFR was 12.67%±10.60% in Stage I specimens, 19.00%±17.89% in Stage II specimens, 16.63%±17.13% in Stage III specimens, and 37.49%±21.1% in Stage IV specimens. The significant difference between Stage IV and Stage II (P=0.001) or Stage III (P=0.001) was identified (Figure 2).
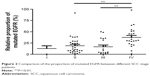  | Figure 2 Comparison of the proportion of mutated EGFR between different SCC stage patients. |
Discussion
In this study, we detected two kinds of deletion and six point mutations in EGFR exon 19 from 89 Chinese male SCC patients. Out of the six point mutations identified, five mutations (M1–M5) are reported, for the first time, to be associated with cancer. In this study, 85% SCC patients were harboring the EGFR mutations; this figure is much higher than that reported previously in other studies (15%–45%).15,31,32 The significant difference may be due to the different methods adopted. In most previous studies, genomic DNA extracted from paraffin-embedded tumor specimens was used to detect mutations.21 The lower concentration of genomic DNA analyzed leads to poor sensitivity and inaccuracy of mutation detection.33
In this study, we used cDNA as the PCR template which was extracted and reverse transcribed from fresh specimens (0.050–0.100 g). The higher concentration of template improved the sensitivity of our system. Additionally, PCR-DGGE is more sensitive (~1%24) than direct sequencing (20%15) and more cost-effective than the high-throughput sequencing method.4 Shahi et al24 used the same PCR-DGGE method and detected 10 different kinds of EGFR exon 19 mutations in 55 out of 210 (16.2%) Caucasian NSCLC patients. Bircan et al32 reported that 45.5% of Turkish male SCC patients (n=11) carried the EGFR exon 19 mutation. These results demonstrate that the EGFR exon 19 mutation rate is significantly different among the ethnic groups.
The EGFR mutation detection and profile are crucial for the determination of the lung cancer treatment strategies. The EGFR mutational profile of NSCLC, especially SCC, is not well studied in the Chinese population.34
In this study, among the EGFR mutation-positive patients, 88.3% of patients (68 of 77) carried the EGFR–TKI-sensitive mutation – deletion 1 (64.9%) and deletion 2 (23.3%). It is an interesting finding because adenocarcinoma patients harboring this mutation can significantly benefit from the EGFR–TKI treatment,30,35 and several studies have demonstrated that the SCC patients could benefit from TKI treatment.17,18
Kaneda et al36 reported that the Del-E746 patients have a better response to EGFR–TKI compared to Del-L747 patients. In 89 Chinese SCC patients assayed in this study, the frequency of the 746-751-deletion and 747-751-deletion was 20.2% and 56.2%, respectively. In 11 Turkish SCC patients, three deletions occurred in the 731-744 regions (27.3%) and two occurred in the 750-760 regions (18.2%).37 These results suggest that more specific EGFR–TKI should be developed for Chinese SCC patients.
In 37 Stage II patients, five kinds of point mutations were detected in 13.5% patients, one kind of point mutation was detected in 22 Stage III patients (13.6%), but no point mutation was identified in 26 Stage IV patients. In other words, in patients harboring the EGFR exon 19 mutation, the frequency of deletion increased from 72.9% (Stage II), 72.7% (Stage III) to 88.4% (Stage IV). The deletion mutation became increasingly dominant with the development of the disease, which agreed with the finding in Caucasian (100%)24 and Turkish SCC patients (100%).32
In this study, one point mutation c.2274A>G was identified in 5 out of 9 patients harboring the point mutation. Though this is a synonymous mutation (Glu (GAA)758Glu (GAG)), GAG (4.39) appears more frequently in high-expressing genes than GAA (2.95).38 Synonymous mutation can play an important role in cancer,39 ie, KRas,40,41 by affecting the gene expression,39,42 especially in oncogenes. The role of new identified point mutation c.2274A>G warrants further study.
Another interesting finding in this study was that the mutated EGFR mRNA in total EGFR mRNA varied from 0.98% to 100%, which possibly explained the discrepancy of EGFR–TKI treatment outcome among EGFR-positive SCC patients.14 In the present study, the proportions of mutated EGFR in total were significantly higher in Stage IV (37.49%) than in Stage II (19.00%) (P=0.001) and Stage III (16.63%) (P=0.001) (Figure 2). Selvaggi et al29 reported that EGFR protein was highly expressed in Stage III (50%) compared to Stage I (20%) and Stage II (25%) in NSCLC patients. Our results further suggest that mutated EGFR protein may be the main contributor to the higher expression of EGFR in advanced-stage SCC or NSCLC, and this issue deserves further attention.
Based on the above-mentioned findings, the EGFR–TKI treatment for Chinese SCC patients and the best time point of the treatment should be reconsidered and studied.
Acknowledgments
This work was supported by grants from the Ministry of Agriculture in China (2012ZX08011002-004), the National Natural Science Foundation of China (31121064), State Key Laboratory of Microbial Metabolism (No 2011DA105494), and State Key Laboratory of Material Sciences.
Disclosure
The authors report no conflicts of interest in this work.
References
Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27(1):2–12. | ||
WHO. Cancer fact sheet No. 297. Reviewed January 20132012. | ||
Fiala O, Pesek M, Finek J, Benesova L, Bortlicek Z, Minarik M. Gene mutations in squamous cell NSCLC: insignificance of EGFR, KRAS and PIK3CA mutations in prediction of EGFR–TKI treatment efficacy. Anticancer Res. 2013;33(4):1705–1711. | ||
Wu K, Huang RS, House L, Cho WC. Next-generation sequencing for lung cancer. Future Oncol. 2013;9(9):1323–1336. | ||
Tetsu O, Hangauer MJ, Phuchareon J, Eisele DW, McCormick F. Drug resistance to EGFR inhibitors in lung cancer. Chemotherapy. 2016;61(S5):223–235. | ||
Genova C, Hirsch FR. Clinical potential of necitumumab in non-small cell lung carcinoma. Onco Targets Ther. 2016;9:5427–5437. | ||
Song Z, Zhang Y. Efficacy of gefitinib or erlotinib in patients with squamous cell lung cancer. Arch Med Sci. 2015;11(1):164–168. | ||
Tan WL, Ng QS. The continuing role of epidermal growth factor receptor tyrosine kinase inhibitors in advanced squamous cell carcinoma of the lung. Transl Lung Cancer Res. 2016;5(1):106–109. | ||
Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;(28 Suppl 1):S24–S31. | ||
Somasundaram A, Socinski MA, Burns TF. Personalized treatment of EGFR mutant and ALK-positive patients in NSCLC. Expert Opinion Pharmacother. 2014;15(18):2693–2708. | ||
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. | ||
Lee VH, Tin VP, Choy TS, et al. Association of exon 19 and 21 EGFR mutation patterns with treatment outcome after first-line tyrosine kinase inhibitor in metastatic non-small-cell lung cancer. J Thorac Oncol. 2013;8(9):1148–1155. | ||
Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res. 2015;5(9):2892. | ||
Fang W, Zhang J, Liang W, et al. Efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors for Chinese patients with squamous cell carcinoma of lung harboring EGFR mutation. J Thorac Dis. 2013;5(5):585–592. | ||
Rossi ED, Gerhard R, Cirnes L, Machado JC, Schmitt F. Detection of common and less frequent EGFR mutations in cytological samples of lung cancer. Acta Cytol. 2014;58(3):275–280. | ||
Chiu CH, Chou TY, Chiang CL, Tsai CM. Should EGFR mutations be tested in advanced lung squamous cell carcinomas to guide frontline treatment? Cancer Chemother Pharmacol. 2014;74(4):661–665. | ||
Xu J, Chu T, Jin B, et al. Epidermal growth factor receptor tyrosine kinase inhibitors in advanced squamous cell lung cancer. Clin Lung Cancer. 2016;17(4):309–314. | ||
Mu XL, Li LY, Zhang XT, et al. Gefitinib-sensitive mutations of the epidermal growth factor receptor tyrosine kinase domain in Chinese patients with non-small cell lung cancer. Clin Cancer Res. 2005;11(12):4289–4294. | ||
Xu J, Zhang Y, Jin B, et al. Efficacy of EGFR tyrosine kinase inhibitors for non-adenocarcinoma lung cancer patients harboring EGFR-sensitizing mutations in China. J Cancer Res Clin Oncol. 2016;142(6):1325–1330. | ||
Hata A, Katakami N, Yoshioka H, et al. How sensitive are epidermal growth factor receptor-tyrosine kinase inhibitors for squamous cell carcinoma of the lung harboring EGFR gene-sensitive mutations? J Thorac Oncol. 2013;8(1):89–95. | ||
Park SH, Ha SY, Lee JI, et al. Epidermal growth factor receptor mutations and the clinical outcome in male smokers with squamous cell carcinoma of lung. J Korean Med Sci. 2009;24(3):448–452. | ||
Kruglyak KM, Lin E, Ong FS. Next-generation sequencing and applications to the diagnosis and treatment of lung cancer. Adv Exp Med Biol. 2016;890:123–136. | ||
de Biase D, Visani M, Malapelle U, et al. Next-generation sequencing of lung cancer EGFR exons 18–21 allows effective molecular diagnosis of small routine samples (cytology and biopsy). PloS One. 2013;8(12):e83607. | ||
Shahi RB, De Brakeleer S, De Greve J, Geers C, In’t Veld P, Teugels E. Detection of EGFR–TK domain-activating mutations in NSCLC with generic PCR-based methods. Appl Immunohistochem Mol Morphol. 2015;23(3):163–171. | ||
Luthra R, Chen H, Roy-Chowdhuri S, Singh RR. Next-generation sequencing in clinical molecular diagnostics of cancer: advantages and challenges. Cancers. 2015;7(4):2023–2036. | ||
Shao D, Lin Y, Liu J, et al. A targeted next-generation sequencing method for identifying clinically relevant mutation profiles in lung adenocarcinoma. Sci Rep. 2016;6:22338. | ||
Gray PN, Dunlop CL, Elliott AM. Not all next generation sequencing diagnostics are created equal: understanding the nuances of solid tumor assay design for somatic mutation detection. Cancers. 2015;7(3):1313–1332. | ||
Khoo C, Rogers T-M, Fellowes A, Bell A, Fox S. Molecular methods for somatic mutation testing in lung adenocarcinoma: EGFR and beyond. Transl Lung Cancer Res. 2015;4(2):126. | ||
Selvaggi G, Novello S, Torri V, et al. Epidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non-small-cell lung cancer. Ann Oncol. 2004;15(1):28–32. | ||
Pallis AG, Voutsina A, Kalikaki A, et al. ‘Classical’ but not ‘other’ mutations of EGFR kinase domain are associated with clinical outcome in gefitinib-treated patients with non-small cell lung cancer. Br J Cancer. 2007;97(11):1560–1566. | ||
Miyamae Y, Shimizu K, Hirato J, et al. Significance of epidermal growth factor receptor gene mutations in squamous cell lung carcinoma. Oncol Rep. 2011;25(4):921. | ||
Bircan S, Baloglu H, Kucukodaci Z, Bircan A. EGFR and KRAS mutations in Turkish non-small cell lung cancer patients: a pilot study. Med Oncol. 2014;31(8):87. | ||
Akbari M, Hansen MD, Halgunset J, Skorpen F, Krokan HE. Low copy number DNA template can render polymerase chain reaction error prone in a sequence-dependent manner. J Mol Diagn. 2005;7(1):36–39. | ||
Qin BM, Chen X, Zhu JD, Pei DQ. Identification of EGFR kinase domain mutations among lung cancer patients in China: implication for targeted cancer therapy. Cell Res. 2005;15(3):212–217. | ||
Zhang X, Chang A. Somatic mutations of the epidermal growth factor receptor and non-small-cell lung cancer. J Med Genet. 2007;44(3):166–172. | ||
Kaneda T, Hata A, Tomioka H, et al. Possible differential EGFR–TKI efficacy among exon 19 deletional locations in EGFR-mutant non-small cell lung cancer. Lung Cancer. 2014;86(2):213–218. | ||
Bircan S, Baloglu H, Kucukodaci Z, Bircan A. EGFR and KRAS mutations in Turkish non-small cell lung cancer patients: a pilot study. Med Oncol. 2014;31(8):87. | ||
Lavner Y, Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene. 2005;345(1):127–138. | ||
Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12(1):32–42. | ||
Lampson BL, Pershing NL, Prinz JA, et al. Rare codons regulate KRas oncogenesis. Curr Biol. 2013;23(1):70–75. | ||
Supek F, Minana B, Valcarcel J, Gabaldon T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156(6):1324–1335. | ||
Hense W, Anderson N, Hutter S, Stephan W, Parsch J, Carlini DB. Experimentally increased codon bias in the Drosophila Adh gene leads to an increase in larval, but not adult, alcohol dehydrogenase activity. Genetics. 2010;184(2):547–555. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

