Back to Journals » Cancer Management and Research » Volume 8
EGFR testing and clinical management of advanced NSCLC: a Galician Lung Cancer Group study (GGCP 048-10)
Authors Vázquez S, Casal J, Afonso Afonso FJ, Fírvida JL, Santomé L, Barón F, Lázaro M, Pena C, Amenedo M, Abdulkader I, González-Arenas C, Fachal L, Vega A
Received 21 March 2015
Accepted for publication 30 August 2015
Published 4 February 2016 Volume 2016:8 Pages 11—20
DOI https://doi.org/10.2147/CMAR.S85173
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Sergio Vázquez,1 Joaquín Casal,2 Francisco Javier Afonso Afonso,3 José Luis Fírvida,4 Lucía Santomé,5 Francisco Barón,6 Martín Lázaro,7 Carolina Pena,7 Margarita Amenedo,8 Ihab Abdulkader,9 Carmen González-Arenas,10 Laura Fachal,11 Ana Vega11
On behalf of the Galician Lung Cancer Group (GGCP)
1Medical Oncology Department, Lucus Augusti University Hospital, Lugo, 2Medical Oncology Department, University Hospital Complex of Vigo, Pontevedra, 3Medical Oncology Department, University Hospital Complex of Ferrol, Ferrol, 4Medical Oncology Department, University Hospital Complex of Ourense, Ourense, 5Medical Oncology Department Povisa Hospital, Vigo, 6Medical Oncology Department, University Hospital Complex of Santiago de Compostela, Santiago de Compostela, 7Medical Oncology Department, Hospital Complex of Pontevedra, Pontevedra, 8Medical Oncology Department, Oncology Center of Galicia, A Coruña, 9Anatomical Pathology Department, University Hospital Complex of Santiago de Compostela, Santiago de Compostela, 10AstraZeneca, Madrid, 11Galician Public Foundation of Genomic Medicine-SERGAS, Santiago de Compostela Clinic Hospital, Santiago de Compostela, Spain
Purpose: This study aimed to assess the incidence of mutations in the epidermal growth factor receptor (EGFR) gene in non-small-cell lung cancer (NSCLC) patients in the Galician region of Spain and the clinical management and outcome of patients carrying EGFR mutations.
Patients and methods: All newly diagnosed advanced or metastatic NSCLC patients were screened for EGFR mutations in matched tumor samples (tissue or cytology specimens) and serum samples.
Results: Of 198 patients screened for EGFR mutations in tumor samples, 184 had evaluable data and, of these, 25 (13.6%) had EGFR mutations (84% sensitizing mutations). EGFR mutation was found in serum in 14 (8.1%) patients (of 174 evaluable). Compared to matched tumor tissue, serum EGFR mutation testing specificity and sensitivity were 99% and 52%, respectively. All but two patients received gefitinib. Median progression-free survival and overall survival were 10 (95% confidence interval: 4.8–15.3) months and 17.8 (95% confidence interval: 13.9–21.6) months, respectively, in patients carrying sensitizing mutations.
Conclusion: The incidence of EGFR mutations in Galicia is consistent with previous data in Spain. Our results also support the feasibility of EGFR testing to guide treatment decision making using tumor tissue or cytology samples, or serum samples if tumor specimens are unavailable. These findings also confirm that first-line gefitinib is an active treatment option in Caucasians with EGFR mutation-positive NSCLC.
Keywords: epidermal growth factor receptor, EGFR tyrosine inhibitors, TKIs, EGFR gene mutation, EGFR mutation testing, non-small-cell lung cancer
Introduction
Non-small-cell lung cancer (NSCLC), accounting for more than 85% of lung cancer cases, is a leading cause of cancer-related death worldwide.1 NSCLC usually presents with advanced stage at diagnosis. Conventional systemic chemotherapy, mainly platinum-based regimens, has been the cornerstone of treatment for advanced NSCLC, although it provides only a modest benefit in survival.2 Increased knowledge of the molecular biology of lung cancer led to the development of the specific anti-EGFR tyrosine kinase inhibitors (TKIs) such as gefitinib and erlotinib. These targeted agents have shown a higher efficacy among patients harboring specific activating mutations in exons 18–21 encoding the tyrosine kinase domain of EGFR gene.3–7 The majority of EGFR activating mutations are exon 19 deletions (45%) and a point mutation (L858R) in exon 21 (40%–45%).8,9 Activating EGFR mutations are significantly more common in Asians, women, never-smokers, and patients with adenocarcinoma histology.5
The enhanced response to EGFR TKIs in patients harboring activating mutations has led major oncology groups to recommend EGFR mutation testing to guide therapeutic decision making.10–12 However, this molecular assay continues to be underused in clinical practice since it is not always feasible. Moreover, despite the variety of methodologies available for EGFR mutation testing,13,14 there is currently no consensus on the optimal detection method. Traditionally, direct sequencing of DNA, especially the Sanger method, has been regarded as the “gold standard” for EGFR mutation testing.15 However, this method is limited by its moderate sensitivity, requirement for high-quality tumor samples, and long turnaround time (TAT).16,17 The drawbacks of direct sequencing and recent advances in molecular techniques have led to the development of commercialized test kits with improved sensitivity and TAT for detecting mutations, such as the Therascreen® EGFR RGQ PCR kit (Quiagen, Manchester, UK).18 Furthermore, sources of tumor material have been another challenging issue in the search for an optimal method for EGFR mutation testing. Formalin-fixed, paraffin-embedded tissue (FFPET) specimens account for the majority of diagnostic samples in clinical practice, although the amount of tumor tissue available for mutation testing is generally very limited. Cytology samples, gathered at diagnosis less invasively than tissue samples, could be an alternative source for mutation testing when tumor tissue samples are not available or have a limited content of tumor DNA.19–21 Recently, the use of surrogate samples such as serum has also attracted attention for mutation testing considering its less invasive collection than other sampling procedures and its nature as a repeatable source. Indeed, the assessment of EGFR mutations in plasma has been demonstrated to be feasible in inoperable NSCLC patients.22
Most of the studies evaluating the predictive role of EGFR mutations in NSCLC were carried out in Japan, where the incidence of EGFR mutations was high (20%–40%). However, data on the incidence of EGFR mutations in Europe, and particularly in Spain, are limited.23,24 Furthermore, the incidence of EGFR mutations could vary among different regions of Spain, given that there is a marked geographical variability of lung cancer,25 mainly based on the distribution of risk factors, such as smoking.
The present study was conducted to assess the incidence of EGFR mutations in newly diagnosed advanced or metastatic NSCLC patients in the Galician region of Spain and the clinical management and outcome of patients with EGFR-positive mutations.
Patients and methods
Study design and patients
This was a multicenter, prospective observational study conducted in nine hospitals in the Galician region of Spain belonging to the Galician Lung Cancer Group (GLCG). The primary endpoint of the study was to estimate the incidence of EGFR-positive mutation in patients with advanced or metastatic NSCLC. Secondary endpoints included the type of EGFR mutations found in EGFR-positive patients, the clinical management of patients carrying EGFR mutations, and the concordance between tumor mutation status and serum samples.
All consecutive adult patients (aged ≥18 years) with histologically or cytologically confirmed newly diagnosed advanced or metastatic NSCLC (stage IIIB noncandidate for locoregional therapy or stage IV) and candidates for first-line therapy were prospectively screened for EGFR mutations during a 13-month period. Patients were further required to have tumor tissue available (taken from primary tumor or metastatic area), obtained fine-needle aspiration (FNA), bronchoalveolar lavage, bronchial brushing, or pleural fluid cytology. Exclusion criteria included histological evidence of mixed small-cell/large-cell carcinoma.
Written informed consent was obtained from all patients before they were included in the study, as was their permission to use their available tumor and serum samples obtained at diagnosis to perform the EGFR mutation analyses. The Independent Ethics Committee of Galicia (SERGAS) approved the study protocol. The study was carried out in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines and applicable regulatory requirements.
EGFR mutation testing was performed in tissue samples (FFPET and fresh tissue specimens) obtained from the original biopsy performed at diagnosis, before any treatment was initiated. In case of lack of tumor tissue, EGFR mutation analysis could be performed in cytology samples obtained at diagnosis. In addition, whenever possible, mutation status was determined in serum samples obtained from any routine blood extraction performed before treatment initiation. All samples used for EGFR mutation testing were required to be obtained through routine procedures carried out under clinical practice conditions.
The therapeutic strategy could be based on the results of EGFR mutation testing; however, physicians’ therapeutic decision making was based on their clinical judgment and clinical practice conditions.
Patients with EGFR-positive mutations were prospectively followed up until premature withdrawal for any cause (including death) or until 9 months from the inclusion of the last patient in the study, whichever came first, in order to characterize their clinical management. EGFR mutational status could be evaluated again in those patients experiencing disease progression who had undergone a biopsy or cytology after disease progression according to routine clinical practice. In addition, whenever possible, EGFR mutation testing was also performed in serum samples after disease progression.
EGFR gene mutation analysis
Genomic DNA extraction and EGFR mutation testing on available tumor samples (biopsy or cytology specimens) was performed at a central laboratory (University Hospital Complex of Santiago de Compostela, Santiago de Compostela, Spain). DNA extraction and EGFR mutation analysis on plasma samples was carried out at the Galician Public Foundation of Genomic Medicine, Santiago de Compostela Clinic Hospital, Santia (Santiago de Compostela, Spain).
EGFR mutation detection in tumor samples was performed using the Therascreen® EGFR RGQ PCR kit (Qiagen), a European Conformity (CE) in vitro diagnostic-marked allele-specific real-time polymerase chain reaction test designed to detect 29 somatic mutations in exons 18, 19, 20, and 21 of the EGFR gene in NSCLC. This certified commercialized test kit is based on mutation-specific amplification achieved using the amplification and refractory mutation system (ARMS) and Scorpions® polymerase chain reaction technologies for detection of amplification.
FFPET and cytology specimen preparation and DNA extraction were previously performed using the DNA FFPE Tissue Kit (Qiagen), following the manufacturer’s instructions. Briefly, genomic DNA was derived from tumor tissue after macrodissection at 4 μm of FFPET blocks and fresh tissue specimens. The tumoral region had previously been located in biopsy sections stained with hematoxylin–eosin or in Diff-Quick or Papanicolaou-stained cytologic preparation. Laser microdissection of the tumor tissue was required in case of samples with low tumor cell content. After paraffin removal (xylene 5 minutes, ethanol 5 minutes), tumor tissue sections were lysed by incubation at 56°C with protein kinase K and binding buffer. Subsequently, isopropanol was added to the lysis mixture and the resulting mixture was centrifuged through a column with a glass fiber filter insert. The adsorbed DNA was washed and then eluted from the column with an aqueous solution. The amount of genomic DNA was quantified by spectrophotometry (NanoDrop ND-1000 instrument; Thermo Fisher Scientific, Waltham, MA, USA) and adjusted to a fixed concentration to be added to the amplification/detection mixture of the Therascreen® EGFR RGQ PCR kit. Target DNA amplification and EGFR mutations detection was performed using the ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). A mutant control and a negative control were used to confirm the validity of each run. Data analysis and reporting of results were fully automated.
Free-circulating DNA from serum was purified using the QIAamp Circulating Nucleic Acid Kit and the QIAvac 24 Plus (both from Qiagen) following the manufacturer’s instructions. Detection of EGFR mutations in serum samples was also carried out using the Therascreen® EGFR RGQ PCR kit. The analysis was performed according to the manufacturer’s instructions, and the reactions were run in the Rotor-Gene Q 5plex HRM instrument and analyzed with the Rotor-Gene Q software (version 2.0.2) (Qiagen). Following the manufacturer’s instructions, Rotor-Gene Q plots from all reactions were checked in order to avoid false positives due to linear increase in fluorescence (which arises due to a fluorescence artifact). After inspection, three samples classified as negative (under the ΔCt cutoff values defined by the manufacturer to identify mutations from FFPET) demonstrated an exponential increase in fluorescence (which is characteristic of true positives). The increase of ΔCt cutoff values by 20% allowed these three samples to be classified as positives. No other changes in mutation status for the remaining samples (N=193) were observed after increasing the ΔCt cutoff values. The three potentially positive samples by serum analyses (after increasing ΔCt cutoff values by 20%) were confirmed as positives when results from FFPET and serum were merged (until then, results from each study were blind to the other).
EGFR mutation status was considered positive when the EGFR mutation testing result was positive in at least one of the samples analyzed in each patient. In the event that one of the samples available was negative for EGFR mutation and the mutational status was unknown for the other one, EGFR mutation status was identified as negative.
Statistical analysis
The TAT was calculated from the date the sample was sent to the laboratory until the results obtained were received by the physicians. The association between EGFR mutation status and clinical characteristics was calculated using the chi-square test or the Fisher’s exact test. EGFR mutation rate between tumor and serum samples was compared by the chi-square test, and the concordance of the mutation rate between tumor (biopsy or cytology) and serum specimens was estimated using Cramer’s V coefficient, where coefficient values closer to 1 indicate a stronger concordance. The sensitivity and specificity of the detection of EGFR mutations in serum specimens with respect to tumor samples were estimated. Positive predictive value (PPV) and negative predictive value were also assessed.
Efficacy analyses were conducted on the patients who had tumors carrying EGFR mutations. Tumor response was evaluated according to the Response Evaluation Criteria In Solid Tumors (RECIST) version 1.1.26 The probability of progression-free survival (PFS) and overall survival (OS) was estimated using the Kaplan–Meier method. PFS was assessed from the start of therapy for NSCLC until documented disease progression or death from any cause. Patients were censored at the date of last follow-up if still alive or without disease progression at the time of the analysis. OS was measured from the start of treatment to death. Patients were censored at the date of last follow-up if still alive at the time of the analysis.
The statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS) version 17.0 (SPSS Inc., Chicago, IL, USA).
Results
Patients
From February 2011 to March 2012, a total of 209 patients from nine Spanish centers were enrolled in the study. Eleven patients were excluded for not fulfilling the inclusion criteria. Therefore, a total of 198 patients were prospectively screened for EGFR mutations. The patient and sample disposition flow diagram is shown in Figure 1. The demographic and clinical characteristics of these patients are shown in Table 1. Briefly, 34% were female, all but one patient was Caucasian, and 22% were never-smokers. The most common histology was adenocarcinoma (69%). Eastern Cooperative Oncology Group (ECOG) Performance Status was 0 or 1 in 68% of patients.
  | Figure 1 Patient and sample disposition flow diagram. |
EGFR mutation status and clinical characteristics
Of the 198 tumor samples available, EGFR mutation analyses were performed on 114 (58%) tumor samples gathered from the biopsy performed at diagnosis (112 from FFPET and two from fresh tissue blocks) and on 84 (42%) cytology specimens (Figure 1). Cytology specimens were mainly obtained through FNA (63%). The source and type of tumor samples for EGFR mutation testing are shown in Table 2.
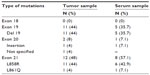
EGFR mutation testing could not be performed in the tumor samples of six (3%) patients due to a too-low content of tumor cells in the tissue sample. In addition, the results of EGFR mutation analysis could not be evaluated in seven (3.5%) patients because of insufficient DNA quality. EGFR mutation data for 184 (93.4%) patients could be assessed (Figure 1). Of these, 25 (13.6%) had EGFR mutations, whereas 159 (86.4%) patients were negative for EGFR mutation. Of the 25 patients with EGFR mutations, 15 (60%) underwent mutation analysis performed on FFPET samples and ten (40%) on cytology specimens. Among patients carrying EGFR mutations, 22 (84%) had sensitizing mutations: 44% had exon 19 deletion and 44% exon 21 L858R point mutation (Table 3). The median TAT was 8 (6–13) days.
  | Table 3 Type of EGFR mutations identified in tumor (N=25) and serum samples (N=14) |
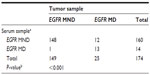
EGFR mutation assessment in serum samples was performed in 196 patients. Of these, 174 patients also had tumor sample results (88.8% of patients with tumor results), of whom 14 (8.1%) had EGFR mutations. Of patients harboring EGFR mutations, seven (50.0%) patients had exon 21 L858R mutation and five (35.7%) patients had exon 19 deletion (Table 3).
There was a significant association between EGFR mutation status in tumor samples and serum specimens (P<0.001). The Cramer’s V coefficient used to assess the overall concordance between the tumor and serum specimens was 0.6. The specificity of serum EGFR mutation testing with respect to tumor mutation status was 99%, whereas the sensitivity was 52%. The PPV and negative predictive value were 93% each (Table 4).
EGFR mutation status between baseline and disease progression could not be performed, given that tumor samples were not obtained at progression due to the refusal of patients to undergo the biopsy or because physicians did not consider the biopsy necessary.
EGFR mutations were more frequently detected in female sex (68%) (P<0.001), patients who had never smoked (68%) (P<0.001), and those with an adenocarcinoma (100%) (P<0.01) (Table 1). Histological confirmation (biopsy) of NSCLC had been performed in 16 (64%) patients, whereas in ten (40%) patients, the diagnosis was confirmed by cytology (FNA). All but one patient had stage IV NSCLC. Local/regional (40%) and pleural (40%) metastases followed by bone (24%) and brain (20%) metastases were mainly found in patients with EGFR mutations.
Treatment
The mean time from diagnosis of advanced or metastatic disease to treatment initiation was 1.2±0.5 months. All but two patients received first-line treatment and the two remaining patients were given best supportive care after mutation testing results were available. Gefitinib was administered as monotherapy in 21 (87.5%) patients and in combination with carboplatin in one patient. Only one patient received chemotherapy plus biological therapy (cisplatin + docetaxel + bevacizumab).
Among patients who received gefitinib, ten (47.6%) patients had tumors containing EGFR mutations in exon 21, nine (42.9%) in exon 19, and two (9.5%) in exon 20. Gefitinib was administered in combination with carboplatin in one patient harboring an EGFR mutation in exon 19 and chemotherapy plus bevacizumab was given to one patient with an exon 21 mutation.
Efficacy
Efficacy analyses were conducted in those patients harboring EGFR mutations who had received first-line therapy (n=23). Three patients were lost to follow-up; therefore, a total of 20 patients had available data for efficacy evaluation.
The overall response rate (ORR) was 60.9%. Partial response was attained in 14 (60.9%) patients, disease stabilization was achieved in two patients (8.7%), and four patients (17.4%) showed disease progression during treatment. The ORR for those patients with EGFR-sensitizing mutations (exon 19 deletion and exon 21 L858R mutation) who received gefitinib (n=17) was 70.6%. Among these patients, two (11.8%) reached disease stabilization (≥12 weeks) and three had progressive disease.
The median duration of follow-up was 9.7 (range: 4.5–15.8) months. The median PFS was 9.7 (95% confidence interval [CI]: 4.0–15.5) months for patients receiving first-line treatment (n=20) and the 1-year PFS rate was 37% (Figure 2A). The median PFS was 9.7 (95% CI: 4.3–15.1) months for patients treated with gefitinib monotherapy (n=18) and 10.0 (95% CI: 4.8–15.3) months for those patients receiving gefitinib monotherapy and carrying EGFR-sensitizing mutations (n=17). The median OS was 17.8 (95% CI: 7.3–28.3) months and the 1-year survival rate was 63% (Figure 2B). The median OS for those patients harboring EGFR-sensitizing mutations who received gefitinib monotherapy was 17.8 (13.9–21.6) months. At the time of the analysis, nearly half of the patients (12 out of 25) were alive. Half of the patients who died harbored EGFR mutations in exon 19, five patients carried a mutation in exon 21, and the remaining two patients had a mutation in exon 20. Fifteen (60%) patients withdrew prematurely from the study, mainly due to death (67%).
Therapy after disease progression
Of the 23 patients who received first-line treatment, eight (34.8%) patients were administered second-line chemotherapy after disease progression. Second-line regimens included pemetrexed, used as monotherapy (four patients) or in combination with cisplatin (three patients), and erlotinib (one patient).
Discussion
The present study found that 13.6% of newly diagnosed patients with advanced NSCLC in the Galician region of Spain carried EGFR mutations. Moreover, our data support the efficacy and safety of first-line gefitinib in NSCLC patients with EGFR mutation-positive tumors.
The frequency of EGFR mutation in our study is consistent with that recently reported in a large European study in a Caucasian population, including Spanish patients with advanced NSCLC (13.7%), wherein the Therascreen® EGFR RGQ PCR kit was also used.23 Furthermore, the EGFR mutation rate was also similar to that detected using the same kit (14%) in formalin-fixed, paraffin-embedded tumor samples from Spanish patients diagnosed with NSCLC.13 However, the mutation rate was lower than that described in the largest Spanish screening for EGFR mutations in NSCLC (16.6%),24 where the incidence of EGFR mutations may have been overestimated, possibly due to the enriched population of women and never-smokers. Additionally, the mutation testing assay applied in this large Spanish study was different from that used in our screening. More recently, a large study conducted in Spain has pointed to an EGFR mutation rate of 11.6%, where different mutation analysis methodologies were used for EGFR mutation testing.27 Despite the differences in sample size, baseline population characteristics, and methodologies used for mutation testing between these national studies and the study presented here, it must be taken into account that this is a regional screening. Hence, the geographical variability in the distribution of risk factors such as smoking or sex may influence EGFR mutation rate. We found that EGFR mutations were more frequent in women and never-smokers, in line with previous reports in Caucasians.13,23
EGFR mutation testing was feasible in the majority of patients (95%) evaluated in our study. This finding supports the idea that EGFR mutation testing should become a routine procedure to guide therapeutic decision making by physicians in clinical practice. Indeed, major oncology groups, including the Spanish Society of Medical Oncology (SEOM) and the Spanish Society of Pathology (SEAP), have recently recommended EGFR mutation testing before treatment initiation.10–12,28
In the current investigation, we found that EGFR mutation testing was feasible in a similar proportion of tissue and cytology samples using the Therascreen® EGFR RGQ PCR kit. Additionally, the EGFR mutation detection rate using this test in cytology samples was comparable to that detected in conventional tumor tissue samples. Our results therefore further strengthen the argument that the use of cytology samples (including FNA and pleural effusion) is an appropriate alternative for EGFR mutation testing when tumor tissue samples are not available or have a low content of tumor DNA.20,21 Indeed, the suitability of cytology samples for routine clinical practice has been recognized in recently published recommendations for EGFR mutation testing.16
In an attempt to assess the convenience of serum samples for EGFR mutation testing, we found that EGFR mutation rates were higher in tumor tissue (13.6% in evaluable samples) than in serum (8.1%) using the Therascreen® EGFR RGQ PCR kit. Compared with matched tumor tissue, the sensitivity and specificity of EGFR mutation tests were 52% and 99% for serum samples. Our results are therefore consistent with those recently reported in Caucasian patients (specificity 99.8% and sensitivity 65.7%) using, as in the study reported here, the Therascreen® EGFR mutation kit.23 Though the moderate sensitivity of EGFR testing seen in serum could recommend against the use of serum samples rather than tissue specimens, the significant concordance and the high specificity and PPV could point to the use of serum as a suitable option in patients when tumor samples are unavailable. However, although these results, together with previous reports suggesting the convenience of serum samples,29–31 are promising, tumor samples should be considered the preferred sample source when available until there is evidence for a higher mutation detection rate using a specific kit for serum samples.
In addition to the requirement for a small amount of testing material, an optimal diagnostic test should offer an accurate result in as short a time as possible to avoid delays in treatment initiation. The median TAT of 8 days to establish the EGFR mutation status using the commercial Therascreen® kit would allow physicians to have available information for treatment individualization in a very short period of time after diagnosis. This is an important concern, as the majority of patients with NSCLC present with advanced or metastatic disease at diagnosis and therefore require prompt initiation of targeted therapy.
More than 90% of patients received first-line treatment with TKIs after the results of EGFR mutation status were available. All of them received gefitinib since it was the only EGFR TKI approved in Europe at the time when the study was conducted. As a descriptive comparison only, the PFS achieved in this study (9.7 months) was comparable to that previously described in the clinical trials carried out with gefitinib in Asian patients with advanced NSCLC.3,32 Moreover, our results are consistent with the PFS (9.7 months) and the OS (19.2 months) data reported in the only study of first-line gefitinib conducted in the European population of Caucasian patients with advanced NSCLC harboring EGFR mutations.23 The ORR achieved in our EGFR mutation-positive NSCLC population (70.6%) is comparable to that previously shown in Asian patients carrying EGFR mutations treated with gefitinib (62% and 74%)3–5 and similar to that described in Caucasian patients harboring activating sensitizing EGFR mutations (69.8%).23 In summary, taken together with recent findings,23 the efficacy results of the current study suggest the benefit in terms of response and survival of first-line gefitinib in Caucasian patients.
The interpretation of our data should take into account the limitations of this study arising from its regional nature and the limited sample population. This study is limited to a geographical region in Spain and may not reflect the incidence of EGFR mutation in NSCLC in the general population of the country. However, regional data could provide interesting information about the incidence of EGFR mutations considering the geographical variability of lung cancer in Spain regarding risk factors such as smoking or sex and some environmental factors. Therefore, despite the obvious limitations of the study, our findings, though modest, might provide a welcome addition to the limited data available on EGFR mutation incidence in a Spanish population with advanced NSCLC and on treatment patterns and clinical outcome of patients carrying EGFR mutations.
Conclusion
The incidence of EGFR mutations in Galicia is consistent with the data previously reported in Spain. Moreover, our results confirm that EGFR mutation testing is a feasible diagnostic platform to guide therapeutic decision making in NSCLC patients in routine clinical practice using tumor tissue or cytology samples. It was also found that serum samples could be considered if tumor specimens are unavailable or tumor DNA is insufficient. Finally, these findings support that first-line gefitinib is effective in Caucasian patients with NSCLC carrying EGFR mutations.
Acknowledgments
The authors would like to acknowledge the Galician Lung Cancer Group (GLCG) for its contribution to the study and for sponsoring the study. We also thank Cristina Vidal and Antonio Torres, from Dynamic, for their editorial and medical writing support, funded by AstraZeneca. The authors would also like to thank AstraZeneca SA for supporting the study through the Investigator Sponsored Studies program.
Author contributions
All authors contributed to the conception and design of the study, acquisition of data, or analysis and interpretation of data. S Vázquez, J Casal, F Javier Afonso Afonso, JL Fírvida, L Santomé, F Barón, M Lázaro, C Pena, and M Amenedo collected data; S Vázquez, J Casal, FJ Afonso Afonso, JL Fírvida, L Santomé, F Barón, M Lázaro, C Pena, M Amenedo, I Abdulkader, C González-Arenas, L Fachal, and A Vega contributed to the analysis and the interpretation of data; and S Vázquez, C González-Arenas, I Abdulkader, and A Vega drafted the manuscript. All authors contributed to critically revising the manuscript for important intellectual content and approved the final manuscript.
Disclosure
Carmen González-Arenas belongs to the Medical Department of AstraZeneca. The authors report no other conflicts of interest in this work.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | |
Schiller JH, Harrington D, Belani CP, et al; Eastern Cooperative Oncology Group. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–98. | |
Maemondo M, Inoue A, Kobayashi K, et al; North-East Japan Study Group. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. | |
Mitsudomi T, Morita S, Yatabe Y, et al; West Japan Oncology Group. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–128. | |
Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. | |
Rosell R, Carcereny E, Gervais R, et al; Spanish Lung Cancer Group in collaboration with Groupe Français de Pneumo-Cancérologie and Associazione Italiana Oncologia Toracica. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. | |
Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011; 12(8):735–742. | |
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. | |
Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007; 7(3):169–181. | |
Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9 Suppl 5:S1–S32. | |
Keedy VL, Temin S, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29(15):2121–2127. | |
Pirker R, Herth FJ, Kerr KM, et al; European EGFR Workshop Group. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol. 2010; 5(10):1706–1713. | |
Angulo B, Conde E, Suárez-Gauthier A, et al. A comparison of EGFR mutation testing methods in lung carcinoma: direct sequencing, real-time PCR and immunohistochemistry. PLoS One. 2012;7(8):e43842. | |
Goto K, Satouchi M, Ishii G, et al. An evaluation study of EGFR mutation tests utilized for non-small-cell lung cancer in the diagnostic setting. Ann Oncol. 2012;23(11):2914–2919. | |
Bakker E. Is the DNA sequence the gold standard in genetic testing? Quality of molecular genetic tests assessed. Clin Chem. 2006; 52(4):557–558. | |
Marchetti A, Felicioni L, Buttitta F. Assessing EGFR mutations. N Engl J Med. 2006;354(5):526–528. | |
Querings S, Altmuller J, Ansén S, et al. Benchmarking of mutation diagnostics in clinical lung cancer specimens. PLoS One. 2011; 6(5):e19601. | |
Vallée A, Le Loupp AG, Denis MG. Efficiency of the Therascreen® RGQ PCR kit for the detection of EGFR mutations in non-small cell lung carcinomas. Clin Chim Acta. 2014;429:8–11. | |
da Cunha Santos G, Saieg MA, Geddie W, Leighl N. EGFR gene status in cytological samples of nonsmall cell lung carcinoma: controversies and opportunities. Cancer Cytopathol. 2011;119(2):80–91. | |
Ellison G, Zhu G, Moulis A, Dearden S, Speake G, McCormack R. EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol. 2013;66(2):79–89. | |
Hagiwara K, Kobayashi K. Importance of the cytological samples for the epidermal growth factor receptor gene mutation test for non-small cell lung cancer. Cancer Sci. 2013;104(3):291–297. | |
Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15(6):2076–2084. | |
Douillard JY, Ostoros G, Cobo M, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer. 2014;110(1):55–62. | |
Rosell R, Moran T, Queralt C, et al; Spanish Lung Cancer Group. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–967. | |
Fernández-Rubio A, Ruano-Raviña A. Cribado de Cáncer de Pulmón. Lung cancer screening. [Report of Health Technology Assessment]. Ministerio de Ciencia e Innovación in Spanish [Spanish Ministry of Science and Innovation]. Available from: http://avalia-t.sergas.es/DXerais/289/avalia-t200706Cribado-ca-Pulmon.pdf. Accessed December 23, 2015. Spanish. | |
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. | |
Esteban E, Majem M, Martinez Aguillo M, et al. Prevalence of EGFR mutations in newly diagnosed locally advanced or metastatic non-small cell lung cancer Spanish patients and its association with histological subtypes and clinical features: the Spanish REASON study. Cancer Epidemiol. 2015;39(3):291–297. | |
Garrido P, de Castro J, Concha Á, et al. Guidelines for biomarker testing in advanced non-small-cell lung cancer. A national consensus of the Spanish Society of Medical Oncology (SEOM) and the Spanish Society of Pathology (SEAP). Clin Transl Oncol. 2012;14(5):338–349. | |
Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009; 27(16):2653–2659. | |
Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7(1):115–121. | |
Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12(13):3915–3921. | |
Inoue A, Kobayashi K, Maemondo M, et al; North-East Japan Study Group. Updated overall survival results from a randomized phase III trial comparing gefitinib with carboplatin-paclitaxel for chemo-naïve non-small cell lung cancer with sensitive EGFR gene mutations (NEJ002). Ann Oncol. 2013;24(1):54–59. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.