Back to Journals » Journal of Inflammation Research » Volume 15
Effectiveness of Soluble CTLA-4-Fc in the Inhibition of Bone Marrow T-Cell Activation in Context of Indoleamine 2.3-Dioxygenase (IDO) and CD4+Foxp3+ Treg Induction
Authors Massalska M , Ciechomska M , Kuca-Warnawin E , Burakowski T, Kornatka A, Radzikowska A, Pawlak D, Muz B , Loniewska-Lwowska A, Palucha A, Maldyk P, Maslinski W
Received 27 January 2022
Accepted for publication 6 September 2022
Published 22 December 2022 Volume 2022:15 Pages 6813—6829
DOI https://doi.org/10.2147/JIR.S359775
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Magdalena Massalska,1 Marzena Ciechomska,1 Ewa Kuca-Warnawin,1 Tomasz Burakowski,1 Anna Kornatka,1 Anna Radzikowska,1 Dariusz Pawlak,2 Barbara Muz,3 Adrianna Loniewska-Lwowska,4 Andrzej Palucha,5 Pawel Maldyk,6,7 Wlodzimierz Maslinski1
1Department of Pathophysiology and Immunology, National Institute of Geriatrics, Rheumatology, and Rehabilitation (NIGRiR), Warsaw, 02-637, Poland; 2Department of Pharmacodynamics, Medical University of Bialystok, Bialystok, 15-222, Poland; 3Department of Radiation Oncology, Cancer Biology Division, Washington University School of Medicine, St. Louis, MO, 63108, USA; 4Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, 02-106, Poland; 5Genomed S.A. Ponczowa 12, Warsaw, 02-971, Poland; 6Department of Rheumoorthopaedic Surgery, National Institute of Geriatrics, Rheumatology, and Rehabilitation (NIGRiR), Warsaw, 02-637, Poland; 7Clinical Department of Orthopaedic and Traumatology of Locomotor System, Enfant-Jesus Clinical Hospital, Warsaw, 02-005, Poland
Correspondence: Magdalena Massalska, Department of Pathophysiology and Immunology, National Institute of Geriatrics, Rheumatology, and Rehabilitation, Spartanska 1, Warsaw, 02-637, Poland, Tel/Fax +48 22 670 94 94, Email [email protected]
Background: Rheumatoid arthritis (RA) is a chronic autoimmune disease with systemic inflammation finally resulting in damaged joints. One of the RA development models suggests bone marrow (BM) as a place of inflammation development further leading to disease progression. We aimed to investigate the potential of CTLA-4-Fc molecule in inducing tolerogenic milieu in BM measured as indoleamine 2,3-dioxygenase (IDO) expression, CD4+Foxp3+ Treg induction, and T cell activation control. The expression of IDO-pathway genes was also examined in monocytes to estimate the tolerogenic potential in the periphery.
Methods: Bone marrow mononuclear cells (BMMC) were stimulated by pro-inflammatory cytokines and CTLA-4-Fc. Next IDO expression, CD4+CD69+ and CD4+Foxp3+ percentage were estimated by PCR and FACS staining, respectively. Enzymatic activity of IDO was confirmed by HPLC in BM plasma and blood plasma. Genes expressed in IDO-pathway were analyzed by NGS in peripheral monocytes isolated from RA patients and healthy controls.
Results: We found that CTLA-4-Fc and IFN-γ stimulation results in IDO production by BMMC. CTLA-4-Fc induced tryptophan catabolism can inhibit mitogen-induced CD4+ T cells activation without influencing CD8+ cells, but did not control CD25 nor Foxp3 expression in BM cells. Significantly higher expression of selected IDO-pathway genes was detected on peripheral monocytes isolated from RA as compared to healthy controls.
Conclusion: This study sheds light on some immunosuppression aspects present or induced in BM. The potential of IDO-mediated pathways were confirmed in the periphery, what may represent the promising candidates for therapeutic strategies in RA.
Keywords: rheumatoid arthritis, bone marrow, CTLA-4-Fc, indoleamine 2,3- dioxygenase, CD4+Foxp3+, monocytes
Introduction
Rheumatoid arthritis (RA) is understood today as a chronic immune-mediated disease with multiple immune cell types and signaling network malfunctions that result in the defective tissue repair process, finally leading to organ damage predominantly in the joints but also in the lungs and vascular system.1 Among factors contributing to RA can be found: the presence of autoantibodies and pro-inflammatory cytokines, altered metabolism, specific microbiome, epigenetic modifications and dysregulated NF-κB signaling.2,3 On the contrast, osteoarthritis (OA) is the degenerative joint disease, being primarily not an inflammatory process and affecting joints but not internal organs.4 The average prevalence of RA is estimated at 0.5–1.0% globally, whereas genetic factors account for 60% of the risk of developing RA.5 Hip OA prevalence is higher in Caucasian population (3–6%) as compared with 1% or less in Asians, Blacks, East Indians or native Americans with age, sex, obesity and genetics being the general risk factors.4 In this study, we investigated the bone marrow microenvironment in context of tolerance induction. We analyzed the two joint diseases with different etiology in order to investigate the influence of settled inflammatory processes on bone marrow tolerance potential.
Since few years, bone marrow (BM) has been considered as an active participant in rheumatoid arthritis pathogenesis. BM maintain memory B and T cells6 and can serve as a secondary lymphoid organ, where antigen presentation take place.7 It was shown to be the reservoir of both inflammatory T cells and Treg cells.8,9 Our group has demonstrated that BM actively takes part in RA development by supporting activation of B cells,10 T cells11 and Th1712 cells and by maintaining impaired Treg cells.13 Cytotoxic T lymphocyte antigen-4 (CTLA-4) is an activation-induced molecule on T cells that bind B7.1 (CD80) and B7.2 (CD86) on antigen-presenting cells (APC) with higher affinity than CD28, playing a key role in T-cell-mediated dominant immunological self-tolerance.14 Recombinant, soluble forms of CTLA-4 were shown to induce indoleamine 2.3-dioxygenase (IDO), the cytoplasmic enzyme responsible for catalyzing the first step reaction of tryptophan (Trp) catabolism, namely the conversion of tryptophan to N-formylkynurenine and kynurenine (Supplementary Figure 1).15 The IDO is expressed on circulating monocytes which further differentiate toward dendritic cells (DC) and macrophages and through depletion of Trp from the microenvironment is responsible for down-modulating T-cell activation and proliferation, thus inducing tolerogenic milieu.15–17 Trp is an indispensable amino acid, required by all cells to synthesize proteins. IDO-producing cells are protected from Trp self-starvation by the expression of the tryptophanyl-tRNA-synthetase (TTS) – enzyme responsible for the association of Trp with its specific tRNA needed to protein synthesis.15,18
Abatacept, a CTLA-4-Ig fusion protein composed of the Fc region of IgG1 fused to the extracellular domain of CTLA-4 binds to CD80 and CD86 and finally blocks CD80/86:CD28 co-stimulatory pathway in T cells. In Europe, abatacept is approved for use in the treatment of progressive RA.19
Recently published data, point out that co-stimulation blockade by abatacept has led to noticed improvement in patient’s condition in many diseases with IDO or BM-connected pathogenesis. Abatacept was shown to interfere (through IDO induction) with autoantibody-mediated cytokine production by monocytes, explaining the observed fast anti-inflammatory effect of this drug and preferential efficacy in patients with high-titer of antibodies to citrullinated proteins (ACPA) and rheumatoid factor (RF).20 It also was shown to help in graft-versus-host disease (GVHD) prophylaxis in successful unrelated donor stem cell transplantation for children with severe sickle cell disease21 and to control activation of BM adaptive immunity in patients with type 2 diabetes (T2D).22 All those data seem to suggest that CTLA-4-Fc might be very helpful in conditioning the immune system finally leading to tolerance induction, especially in BM microenvironment.
Monocytes, which are BM-derived leukocytes, are involved in RA progression, however under special conditions may contribute to tolerance induction due to production of IDO.17,23,24 As monocytes and macrophages were proofed to originate from a common BM-derived precursor, we decided to analyze the genes involved in IDO and AHR-mediated pathways in peripheral monocytes to estimate their tolerogenic potential.25,26
In this study, we investigated the influence of CTLA-4-Fc on BM cells in the context of induction of tolerogenic conditions created by IDO and CD4+Foxp3+ regulatory T cells (Treg). We demonstrated that CTLA-4-Fc is able to induce production of enzymatically active IDO, but does not increase the Treg number in BM microenvironment. Using RNA-sequencing (RNA-seq) analysis of total RNA of peripheral monocytes and bioinformatic protein–protein interactions (PPI) analysis we have shown the tolerogenic potential and network between IDO- and AHR-pathway proteins in periphery.
Materials and Methods
Patients and Cells
Bone marrow mononuclear cells (BMMC) were obtained from patients with rheumatoid arthritis (RA) and osteoarthritis (OA) who were undergoing complete hip bone replacement surgery. Femoral bone marrow samples were obtained during this surgery as a standard procedure. Peripheral blood (20 mL) was obtained from 11 RA and 11 OA patients before surgery. Additionally, peripheral blood was obtained from 4 RA and 4 healthy volunteers for monocyte isolation further proceeded in RNA-seq. All patients fulfilled the American College of Rheumatology revised criteria for RA27 and OA.28 The RA patient group was composed of 23 patients (19 females and 4 males, mean age 52.6 ± 2.3, age range 26–73 years). Thirteen RA patients were treated with methotrexate, 7 with sulfasalazine and 3 patients were cured with leflunomide belonging to conventional synthetic disease-modifying antirheumatic drugs (DMARDs). Eighteen RA patients were treated with steroids (9 with encorton and 9 with metypred) and 13 claimed to take non-steroidal anti-inflammatory drugs (NSAIDs). The OA patient group was composed of 17 patients (10 females and 7 males, mean age 57.8 ± 1.9, age range 45–68 years). Erythrocyte sedimentation rate (ESR) in both patient groups was as follows: 35.6 ± 4.1 [mm/h] (range 3–79 mm/h) in RA and 13.1 ± 3.1 [mm/h] (range 3–52 mm/h) in OA. The healthy controls (HC) group investigated in NGS experiments was built by 4 patients (4 females, mean age 43.3 ± 8.0, age range 34–67 years). All clinical data concerning patients together with comorbidities are summarized in Table 1.
 |
Table 1 Patients` Demographic and Clinical Characteristics |
Cell Isolation and Culture Conditions
BMMC and peripheral blood mononuclear cells (PBMC) were isolated, respectively, from human BM or venous blood by Ficoll-Paque PLUS (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) density centrifugation. BM samples were diluted four times in PBS (Sigma, St Louis, Missouri, USA) with sodium citrate as an anticoagulant before cell isolation.11 Blood samples were not diluted. BM and blood plasma samples were obtained by centrifugation by 1000×g. Isolated BMMC and PBMC were further used in RT-PCR analysis of chosen genes or in functional assay culture with phenotype assessment. BMMCs or PBMCs were cultured in 24-wells culture flasks (1×106 cells/1mL/well) (Nunc, Roskilde, Denmark) in RPMI-1640 (Invitrogen, Paisley, UK), supplemented with l-glutamine (2 mM), HEPES (10 mM), penicillin (100 IU/mL), streptomycin (100 μg/mL), kanamycin (100 μg/mL), plasmocin (25 μg/mL; InvivoGen, San Diego, CA, USA), and 10% fetal calf serum (FCS) (Biochrom AG, Berlin, Germany) for 12–48 h, according to experiment setting. Monocytes for RNA-seq analysis were isolated from PBMC of 4 RA patients and 4 HC according to the manufacturer’s protocol with the CD14+ MACS bead isolation kit (Miltenyi-Biotec, the Netherlands) as previously described.23 RNA from CD14+ monocytes was isolated using miRNeasy kit according to the manufacturer’s protocol (Qiagen, Manchester, UK). Following isolation, the quality and integrity of the total RNA of all samples (4 RA and 4 HC) were assessed with Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA).
Treatment of Cells with CTLA-4-Fc or Pro-Inflammatory Cytokines and Quantification of IDO Transcripts Using RT-PCR
Freshly isolated BMMC (1x106 cells/1mL/well) were incubated with CTLA-4-Fc (1 or 4 μg/mL) or one of a few pro-inflammatory cytokines (IFN-γ, IL-15, IL-17, TNF, IL-6, IL-1β). Cultures were initiated with: 0.1–10 ng/mL IFN- γ, 10 ng/mL IL-15, 10 ng/mL IL-17, 10 ng/mL TNF, 5 ng/mL IL-6 or 1 ng/mL IL-1β (all from R&D Systems, Minneapolis, MN, USA). After 12h incubation, total RNA was extracted from the cells and IDO together with GAPDH expression were analyzed by RT-PCR. Total cellular RNA extraction, cDNA template preparation and the RT-PCR were done as previously described.29 The PCR products were fractionated by 1.5% agarose gel electrophoresis and densitometrically scanned using Kodak1D Image analysis software (Eastman Kodak, Rochester, NY). Results are presented as ratios between the target gene mRNA and the GAPDH mRNA.
Kinetics of IDO, TTS, IFN-γ, IL-15 and IL-15R Expression by BMMC
To analyze the kinetics of IDO, TTS, IFN-γ, IL-15, IL-15R and GAPDH mRNA expression, BMMC (1x106 cells/1mL/well) were stimulated with 4 μg/mL rh CTLA-4-Fc (R&D Systems, Minneapolis, MN, USA) for 4–24 hours. The high-affinity IL-15R is heterotrimeric composed of β, common γ-chain and IL-15-specific IL-15Rα chain. Here, we investigated expression of IL-15Rα chain that has two isoforms R1 (444pb) and R2 (543bp). There are also two forms of IL-15 detectable: 409 bp and 528 bp. At the time of cell culture termination, total RNA was extracted from the cells and gene expression in stimulated and non-stimulated cells (control) was analyzed by RT-PCR as described above. The sense and antisense primers and lengths of the PCR products of the investigated genes are shown in Table 2.
 |
Table 2 Sequences of Used Primers and Expected Product Sizes |
CD4+ and CD8+ T-Cell Activation Assay by PHA
BMMC were cultured in 24-well plates (1×106 cells/1mL/well) in the absence or presence of CTLA-4-Fc (4 μg/mL) for 48h according to Boasso et al.15 Additionally, some cultures were set in the presence of the IDO inhibitor, 1-methyl-DL-tryptophan (1-MT; final concentration: 1 mM; Sigma-Aldrich, St. Louis, MO, USA) to test if the CTLA-4-Fc effect in T-cell activation is dependent on Trp catabolism. After 48h of culture, T-cell activation was obtained by the addition of phytohemagglutinin A (PHA; Sigma-Aldrich, St. Louis, MO, USA) at a final concentration of 1μg/mL. Generally, CD69 expression was checked on gated CD4+T cells and CD8+ T cells in four different conditions: (control) – no pre-treatment (for 48h), no activation (for 24h); (PHA) – no pre-treatment (for 48h), subsequent PHA activation (for 24h); (CTLA-4-Fc+PHA) - CTLA-4-Fc pre-treatment (for 48h), subsequent PHA activation (for 24h); (CTLA-4-Fc+1-MT+PHA) - CTLA-4-Fc+1-MT pre-treatment (for 48h), subsequent PHA activation (for 24h). After 24h activation with PHA, cells were prepared for cytofluorometric staining of membrane CD3, CD4, CD8, CD69 antigens.
Membrane CD3, CD4, CD8 and CD69 Detection by Flow Cytometry
To examine the phenotype of PHA-activated T cells, the cells were washed with FACS Buffer (BD Biosciences) and membrane antigen expression was determined by incubating cells for 30 minutes in 4֯C in darkness with the following mouse anti-human antibodies: anti-CD3-PE (1.5 μg/mL), anti-CD4-FITC (50 μg/mL), anti-CD8-PerCP (6.25 μg/mL), anti-CD69-APC (50 μg/mL) or with control, isotype matched antibodies. All antibodies were mouse IgG1 isotype and from BD Biosciences, San Jose, CA, USA. Cells were acquired using FACSCalibur flow cytometer (BD Biosciences), and the results were analyzed using CellQuest (BD Biosciences) software. The gating strategy was based on the identification of lymphocytes according to FSC and SSC signal distribution. Then, CD4+ or CD8+ lymphocytes were gated based on CD3 together with CD4 or CD8 expression followed by CD69 expression analysis. Gates were settled according to the isotype control for the desired marker and a minimum of 10,000 cell events were acquired for each sample.
Anti-CD3 T-Cell Activation Assay
In order to investigate the potential of CTLA-4-Fc to induce Treg phenotype in BM CD25− T cells, we performed experiments similar to Razmara et al.30 Cells were treated with CTLA-4-Fc (4 μg/mL) for 1h, prior to stimulation with anti-CD3 mAb (clone Hit3a; 2.5 μg/mL, BD Pharmingen, San Diego, CA, USA). After 2 days in culture, cells were washed and cultured for another 2 days before harvesting and cytofluorometric staining of membrane (CD4, CD25, CD69) and intracellular (Foxp3) antigens.
Foxp3 Detection by Flow Cytometry
To examine the phenotype of anti-CD3-activated T cells, including regulatory T cells (Tregs), the following antibodies were used for detection of membrane antigens: anti-CD4-FITC (50 μg/mL), anti-CD25-PE (12.5 μg/mL), anti-CD69-APC (50 μg/mL) by incubating cells for 30 minutes in 4֯C in darkness. All antibodies were mouse IgG1 isotype and from BD Biosciences, San Jose, CA, USA. Next, cells were fixed and permeabilized using Foxp3 Staining Buffer Set, followed by intracellular staining with anti-Foxp3-PeCy7 antibody (rat IgG2a isotype; 25 μg/mL) according to the manufacturer protocol (PCH101; eBioscience, San Diego, CA, USA). In all variants of the experiment, cells were also stained with control, isotype matched antibodies. Cells were acquired using Canto II flow cytometer (BD Biosciences), and the results were analyzed using Diva (BD Biosciences) software. The gating strategy was based on the identification of lymphocytes according to FSC and SSC signal distribution. Then, CD4+ lymphocytes were gated; followed by the identification of CD4+Foxp3+ cells. Gates were settled according to the isotype control and a minimum of 10,000 cell events were acquired for each sample.
High-Pressure Liquid Chromatography (HPLC) Determination of Kynurenine
IDO functional activity was measured in BM plasma or blood plasma in terms of PBMC and BMMC ability to metabolize tryptophan to kynurenine measured by HPLC as previously reported.31
Heatmap of Selected Gene Expression of IDO- and AHR-Mediated Pathways and Construction of Protein-Protein Interaction (PPI) Network and Modular Analysis
High-throughput next-generation sequencing (NGS) was carried out using TrueSeq Stranded Total RNA Library Prep kit (Cat. No RS-122-2201; Illumina, USA) for 8 profiles (4 HC and 4 RA) of total RNA, according to the manufacturer protocol.23 Differentially expressed genes (DEGs) involved in IDO and AHR-mediated pathways were mapped onto the STRING database to estimate protein–protein interactions (PPI).32
Statistical Analysis
The differences within the groups of patients were tested for their statistical significance using parametric two-tailed T-test or Wilcoxon test when the normality assumption was not met. Comparison of BM plasma with blood plasma from the same patients (done separately for OA and RA patients) were analyzed by paired test t, while comparison of variants of the culture within OA and RA patient groups was analyzed by Wilcoxon test. Data were analyzed using Statistica (version 6.0) and Prism 4.0 (GraphPad, La Jolla, USA) software. For all tests, a value of p < 0.05 was considered significant.
Results
IFN-γ and CTLA-4-Fc-Triggered IDO Expression by BMMC
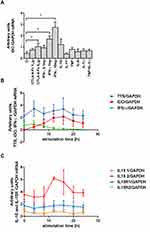
We tested the effect of a few pro-inflammatory cytokines present in RA BM and CTLA-4-Fc on IDO expression by BMMC. Stimulation of BMMC by recombinant human CTLA-4-Fc fusion protein (4 μg/mL) resulted in significant induction of IDO expression (1.05 ± 0.28 vs 0.43 ± 0.16; p = 0.03). Among tested cytokines, only IFN-γ induced effectively IDO in comparison to control in a dose-dependent manner (1.7 ± 0.28 vs 0.43 ± 0.16 p = 0.03 and 2.74 ± 0.47 vs 0.43 ± 0.16 p = 0.03 for 1 ng/mL and 10ng/mL of IFN-γ respectively). IL-17, TNF, IL-6, IL-1β did not induce IDO expression, while stimulation by IL-15 resulted in increased IDO expression, although the results were not statistically significant (Figure 1A).
Next, we investigated the kinetics of CTLA-4-Fc-induced enzymes involved in Trp catabolism together with IL-15 and IL-15R, as IL-15 is proved to be involved in RA pathogenesis and is widely expressed in BM.11,33 The results, presented in Figure 1B and Figure 1C, indicate that: (1) CTLA-4-Fc induced increase in IDO mRNA expression after 12 and 16 hours compared with untreated cells; (2) TTS expression was on constant and high level independently from the time of stimulation; (3) IFN-γ mRNA was higher in CTLA-4-Fc-treated BMMC than in the untreated cells only at 4 and 8 hours of stimulation. CTLA-4-Fc stimulation increased the expression of one of the IL-15 isoforms (length 528 bp), but not another IL-15 isoform (length 409 bp) nor both IL-15Rα isoforms. The increased IL-15 production in BM support the lymphocytes T activation in the BM microenvironment, what may counteract the regulatory conditions created by IDO resulted from CTLA-4-Fc stimulation.11 Further investigation would be needed to verify this hypothesis. The BMMC may produce IDO after stimulation by CTLA-4-Fc or IFN-γ and constant expression of TTS makes the BM cells prepared for effective IDO production without the risk of self-starvation from lack of Trp.
PHA-Triggered Activation of CD4+Cells Prevented by Tryptophan Catabolism
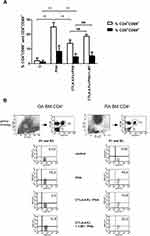
IDO activity is reported to be associated with the blocking properties of T-cell activation and proliferation.34,35 Here, we primed BMMC with CTLA-4-Fc followed by the surface expression of the T-cell activation marker CD69 on CD4+ and CD8+ cells, after stimulation with PHA. There was small percentage of CD4+CD69+ (1.95% ± 1.92%) and CD8+CD69+ (1.18% ± 1.06%) among not activated BMMC (Figure 2A and Figure 2B). Activation with PHA resulted in significant percentage increase of CD4+CD69+ cells (24.95% ± 3.1% vs 1.95% ± 1.92%; p = 0.0078) as well as CD8+CD69+ (8.34% ± 4.0% vs 1.18% ± 1.06%; p = 0.0078). BMMC pretreatment by CTLA-4-Fc for 48h significantly reduced the percentage of activated CD4+ cells (13.83% ± 2.34% vs 24.95% ± 3.1%; p = 0.0078) and CD8+ cells (4.67% ± 2.09% vs 8.34% ± 4.0%; p = 0.0078). In order to test if the inhibitory effect of CTLA-4-Fc was dependent on IDO activation, BMMC pre-incubated with CTLA-4-Fc were in the presence of an inhibitor of IDO, namely 1-MT for 48h before the activation with PHA. Inhibition of IDO activity by 1-MT, prevented the negative effect of CTLA4-Fc on CD4+ T cell activation shown as the percentage of CD4+CD69+, although the results did not meet the statistical significance (18.66% ± 1.35% vs 13.83% ± 2.34%; p = 0.0547). Figure 2A shows results presented for both groups of patients together, Figure 2B presents example staining of OA and RA cells in order to show that the results were not different between the groups of patients. The IDO activity was not reversed by 1-MT in the case of CD8+CD69+ cells. These results suggest that CTLA-4-Fc- induced tryptophan catabolism can inhibit mitogen-induced CD4+ T cell activation without influencing CD8+ cells.
The Effect of CTLA-4-Fc on BM CD4+ T Cells Activation by Anti-CD3
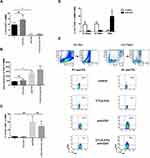
Because we have observed that PHA-triggered activation of T cells manifested as expression of CD69 on CD4+cells is prevented by tryptophan catabolism induced by CTLA-4-Fc treatment, we wanted to investigate if that observation is also valid in case of non-polyclonal activation. Stimulation of BMMC by CTLA-4-Fc increased the expression of CD69 on CD4+ cells, but the results were not statistically significant (Figure 3A). Interestingly, stimulation by anti-CD3 Abs instead of suspected increase of CD4+CD69+ percentage led to statistically significant decrease of CD4+CD69+ percentage as compared to control (1.2% ± 0.36% vs 9.7% ± 2.1%; p = 0.0078; Figure 3A) and stayed unchanged even when cells were pretreated with CTLA-4-Fc. However, the mean expression of CD69 on cells from BM increased after anti-CD3 Abs stimulation (1334.0 ± 216.4 vs 406.6 ± 34.2; p = 0.0078; Figure 3B). What is more, these changes were independent from diagnosis of the patient and different from those observed on paired PBMC. In this case, anti-CD3 stimulation increased the percentage of CD4+CD69+ cells from peripheral blood, while pretreatment with CTLA-4-Fc has lowered it as presented in Figure 3C. The results obtained from peripheral blood did not meet the statistical significance probably because of the small number of probes investigated (n = 4). The comparison of CD69 expression triggered by anti-CD3 Abs (Figure 3D and Figure 3E) observed on CD4+ cells from BM and peripheral blood indicates that indeed one of the features of activation, namely CD69 expression, manifests differently depending on the cell source.
The Effect of CTLA-4-Fc on Percentage of CD4+CD25+and CD4+Foxp3+among BMMC Cells
CTLA-4-Fc was reported to convert naive CD4+CD25− T cells into CD4+CD25+ regulatory T cells in mice.30 Here, anti-CD3 stimulation resulted in statistically significant increase of CD4+CD25+ (66.87% ± 11.3% vs 3.01% ± 0.67%; p = 0.0156) and CD4+Foxp3+ (51.53% ± 11.8% vs 10.66% ± 1.43%; p = 0.0156; Figure 4A-C) percentage among BMMC from OA and RA patients, independently from disease diagnosis. However, CTLA-4-Fc neither alone nor with anti-CD3 Abs was able to influence the BM Treg percentage. The representative plots showing FACS staining for BMMC from OA and RA patients are shown on Figure 4C. Thus, in BM cells, CTLA-4-Fc does not seem to modify the CD4+Foxp3+ Treg compartment at least in the context of CD25 and Foxp3 expression.
IDO Activity in BM and Peripheral Blood
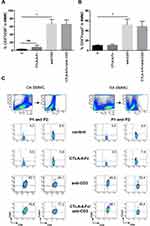
IDO degrades enzymatically tryptophan to kynurenine. To determine the functional activity of IDO expressed in BM and PBMC, we analyzed the kynurenine ratio in blood plasma and BM plasma in paired probes from the same patient. As shown on Figure 5A, kynurenine level was statistically higher in blood than BM (1.54 ± 0.13 vs 0.87 ± 0.09; p = 0.0078 and 1.61 ± 0.08 vs 0.81 ± 0.07; p = 0.03 in RA and OA respectively), what may support the notion of higher tolerance potential in periphery than in BM.
Aryl hydrocarbon receptor (AHR) is claimed to be a crucial factor in the regulation of immune responses, including the IDO role for the development of Treg.36 AHR mediates cellular responses to kynurenine by binding in the nucleus AHR nucleus translocator (ARNT) followed by other genes including cytochromes P450 (CYP).36 As kynurenine analysis has displayed higher activity or concentration of IDO enzyme in peripheral blood, in the last set of experiments, we have performed transcriptomic analyses of selected genes, which are involved in IDO and AHR-pathways using RNA-seq in monocytes from RA patients compared to healthy controls (Figure 5B). Fold change of the main enzymes responsible for tryptophan degradation in IDO-pathway like IDO-1, kynurenine 3-monooxygenase (KMO) or kynurenine aminotransferase (KAT) indicate their higher expression in RA monocytes as compared to HC. From AHR-mediated pathway the expression of the most important proteins, namely AHR and ARNT in RA patients was comparable to HC, what may suggest that it is rather the IDO pathway that limits the suppression induction in the periphery in RA. PIP network analysis supported the existence of 3 nodes interacting with each other in the periphery, namely IDO, CTLA-4 and AHR, giving some data for further investigation (Figure 5C). As there was no difference in expression between RA and HC of few proteins displayed in PIP analysis like estrogen receptor (ESR1), AH receptor-interacting protein (AIP) and Heat shock protein HSP 90-alpha (HSP90AA1) they have been skipped from heatmap presented in Figure 5B.
Discussion
Indoleamine-2,3-dioxygenase (IDO)1 and IDO2 are two closely related tryptophan catabolizing enzymes, induced by inflammatory conditions and contributing to immune responses.37 IDO1 is expressed in immune and non-immune cells, while IDO2 expression is restricted to the liver, kidney and antigen-presenting cells.38 IDO1 immunoregulatory role in mediating tumor immune evasion has been well investigated in human tumors and mouse tumor models.39,40 This results not only from induction of regulatory T cells, but also from the fact that IDO1 is a mediator of inflammatory neovascularization in some tumor development.41 Despite the proved role in cancer control, described effects of IDO1 in autoimmune responses are contradictory.42–45 IDO2 has been much less studied than IDO1. Its activity is required for Treg development in the same way as IDO1, but its role in tumor development control is ambiguous (pro- and anti- inflammatory). IDO1 has robust Trp catabolizing activity, while IDO2`s enzymatic activity is very weak and thus its main function might not mediate through Trp catabolism but through an as yet unidentified pathway, probably supporting autoreactive B cell responses.37
IDO/kynurenine pathway is believed to be a major regulator of the immune system, involved in several central and peripheral disorders including musculoskeletal system as well as central nervous system disorders.46 We have shown that BMMC can express two functionally active enzymes involved in Trp metabolism: IDO and TTS. It is known that the decreased concentration of Trp in the microenvironment affects T cell activity but not the IDO-expressing cells themselves, thus BMMC seems to be protected from negative effects of Trp depletion similarly to “professional” IDO producers.15 Among various pro-inflammatory cytokines present in RA, only IFN-γ stimulated IDO expression in BMMC, which is compatible with data obtained by others.15,47 These results might be valuable not only because of possible tolerance induction in BM, but also in the context of recently published results demonstrating evidence that IFN-γ induces the entry of tumor-repopulating cells (TRCs), being self-renewing stem-like cancer cells, into dormancy through the IDO-Kyn-AHR-p27 cascade.48 IFN-γ is known as a crucial anti-tumor immune factor triggering the JAK/STAT signaling cascade that leads to tumor cell death. On the other hand, IFN-γ upregulates IDO, which facilitates tumor escape from immune elimination and play a role in tumor equilibrium, when growth of tumors cells are controlled by immune system. Results published by Liu et al provide a new direction to develop novel treatment strategies for preventing tumor resistance and recurrence.48
Overexpression of IDO results in blocking T-cell activation and proliferation.34 We have shown that although PHA induced statistically significant increase in the percentage of BM CD4+CD69+ as well as CD8+CD69+, the activation of CD8+ cells was much lower than CD4+ cells. CD8+ cells were also much less affected by CTLA-4-Fc pretreatment, suggesting that CD4+ cells might be more susceptible for tryptophan starvation than CD8+ cells. Similar results were observed on CD4+ and CD8+ cells from peripheral blood, where CD8+ cells were shown to be the only cell population from PBMC with low IDO and high TTS expression, pointing to special protection of this population.15 Surprisingly, polyclonal stimulation by anti-CD3 Abs significantly decreased the percentage (but not mean expression) of BM CD4+ cells expressing CD69. Interestingly, quite opposite results were observed on the same population isolated from PBMC, where anti-CD3 stimulation resulted in the increased number of CD4+CD69+ cells (lack of statistical significance in that case resulted from low patient numbers analyzed). CD69 is best suited for assessing activation induced by polyclonal activators, however it is rapidly expressed after activation (less than 4 h) and also quickly (by 72h of activation) reduced to near background level.49 Basal expression of CD69 is increased on CD3+CD4+ and CD3+CD8+ T cells from RA BM, but it is hard to explain why the percentage of CD69+ cells decreases below the control level after anti-CD3 stimulation.11
The generation of natural Treg cells is highly dependent on IL-2 availability, while the development of adaptive Treg cells can be achieved by several conditions like cytokines (TGF-β, IL-4, IL-10, IL-13) or contact with the certain kind of DC.50 CTLA-4-Fc stimulation together with TCR ligation resulted in induction of Treg phenotype in cells isolated from CD25− mice, as shown by Razmara et al.30 However, we were not able to support those results by our data. In our hands stimulation by CTLA-4-Fc and anti-CD3 did not change the percentage of Foxp3+ cells obtained by anti-CD3 stimulation as shown on Figure 4B and Figure 4C. IDO is known to induce the regulatory phenotype in naive T cells, but we did not have such observations, although we have shown that enzymatically active IDO is produced by BMMC.50 Strong TCR stimulation (like anti-CD3 Ab) was shown to be indispensable for Treg induction as well as stabilization of Foxp3 expression and we have shown that this mechanism is valid also for Treg phenotype induction in BM.51 The requirements for exogenous IL-2 needed for expansion and survival are higher in natural Treg cell populations than in adaptive Tregs. It is not known so far if Tregs present in BM belong mostly to natural or adaptive population.13 Because CTLA-4 as a co-stimulation blocker interferes with IL-2 production, it is possible that stimulation by this compound might result more in inhibition of natural Tregs than adaptive Tregs population. As the number of Tregs in BM did not change in the presence of CTLA-4-Fc, this suggests that Tregs present in BM are mostly adaptive ones, not so sensitive to IL-2 concentration. Another conception is that DC present in BM are less mature phenotypically with lower expression of B7-2, similarly to those from non-obese diabetic (NOD) mice, an animal model for type 1 diabetes, and thus unable to trigger Treg promoting phenotype.52 The reduction of T cells expressing CD25 and Foxp3 was shown in RA patients following abatacept, what suggest Tregs propagation dependence on CD28 signals.53,54 Interestingly, regulatory functions of Tregs after abatacept treatment were enhanced as presented by Alvarez-Quiroga et al.53
IDO activity is mirrored by levels of kynurenine metabolites, easily detected in body fluids like synovial fluid or blood serum. Previously, first investigations of the kynurenine levels in RA patients have shown reduced concentration of tryptophan correlating with disease activity as compared to healthy controls,55 but later data did not support those observations.56 We have shown for the first time significantly higher kynurenine levels in the blood serum than BM serum in RA patients, which suggests more pronounced tolerance induction in the periphery than in BM. Knowing that IDO expressing cells (macrophages and DC) differentiate from monocytes, we performed transcriptomic analysis of IDO and AHR genes on monocytes from RA and HC, which has shown higher expression of genes involved in IDO pathway in peripheral monocytes from RA patients than HC. These data might suggest that chronic illness, which is RA, induced attempts to constrain this process by inducing tolerance milieu through IDO. Grohmann et al have shown that Tregs can trigger high levels of IDO expression in mice DCs through binding of CTLA-4 on Tregs to B7-1 (CD80) and B7-2 (CD86) on DCs.57 Interestingly, NGS analysis has shown that there was no difference in expression of CD80 and CD86 on monocytes between RA and HC, which may suggest that potential for tolerance induction by Treg is similar in RA and HC. Tregs are believed to be a key regulator of tolerance and immunity with IDO being one of the effector mechanisms of Tregs.
All presented above data support the concept of bone marrow being not only a secondary lymphoid organ and place of antigen presentation, but also a place of tolerance induction. These facts might be helpful in developing new therapeutic strategies not only for RA, but generally for bone marrow connected diseases.
Conclusion
These data broaden our knowledge about possible anti-rheumatic effects of CTLA-4-Fc transferred to BM. Fact that enzymatically active IDO can be produced in BM and that genes involved in the IDO/kynurenine pathway are highly expressed in peripheral RA monocytes might be helpful in the context of novel therapies, presently developed and based on tolerance induction.
Abbreviations
ACPA, antibodies to citrullinated proteins; AIP, AHR-interacting protein; AHR, aryl hydrocarbon receptor; APC, antigen-presenting cells; ARNT, AHR nucleus translocator; BM, bone marrow; BMI, body mass index; BMMC, bone marrow mononuclear cells; csDMARDs, conventional synthetic disease-modifying antirheumatic drugs; CTLA-4, cytotoxic T lymphocyte antigen-4; CYP, cytochromes P450; DC, dendritic cells; ESR, erythrocyte sedimentation rate; ESR1, estrogen receptor; FSC, forward scatter; GVHD, graft-versus-host disease; HC, healthy controls; HSP90AA1, heat shock protein (HSP 90)-alpha; IDO, indoleamine 2.3-dioxygenase;IFN-γ, interferon gamma; IL-15, interleukin-15; IL-15R, interleukin-15 receptor; KAT, kynurenine aminotransferase; KMO, kynurenine 3-monooxygenase; MSC, mesenchymal stem cells; 1-MT, 1-methyl-DL-tryptophan; NSAIDs, non-steroidal anti-inflammatory drugs; OA, osteoarthritis; PBMC, peripheral blood mononuclear cells; PPI, bioinformatic protein–protein interactions; RA, rheumatoid arthritis; RNA-seq, RNA-sequencing; SSC, side scatter; RF, rheumatoid factor; T2D, type 2 diabetes; Treg, CD4+Foxp3+ regulatory T cells; TRCs, tumor-repopulating cells; Trp, tryptophan; TTS, tryptophanyl-tRNA-synthetase.
Ethics Approval and Informed Consent
All participants gave a written informed consent according to the Declaration of Helsinki, and the study was approved by the Institute of Rheumatology (now the National Institute of Geriatrics, Rheumatology, and Rehabilitation) Ethics Committee in Warsaw with decisions dated 22.04.2004, 27.06.2014, and KBT 14/1/2016.
Acknowledgments
We are very grateful to Dr Leszek Roszkowski for the help in patient data collection and to Malgorzata Manczak for the help in statistical analysis interpretation.
Authors’ Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the core grant No S/9 National Institute of Geriatrics, Rheumatology, and Rehabilitation from the Polish Ministry of Science and Higher Education and grant No 2018/30/E/NZ5/00104 from the National Science Centre (M.C.).
Disclosure
The authors declare no conflict of interest.
References
1. Weyand CM, Goronzy JJ. The immunology of rheumatoid arthritis. Nat Immunol. 2021;22(1):10–18. doi:10.1038/s41590-020-00816-x
2. Firestein GS, McInnes IB. Immunopathogenesis of rheumatoid arthritis. Immunity. 2017;46(2):183–196. doi:10.1016/j.immuni.2017.02.006
3. Roberti A, Chaffey LE, Greaves DR. NF-kappaB signaling and inflammation-drug repurposing to treat inflammatory disorders? Biology. 2022;11(3):372. doi:10.3390/biology11030372
4. Lespasio MJ, Sultan AA, Piuzzi NS, et al. Hip osteoarthritis: a primer. Perm J. 2018;22:17–84. doi:10.7812/TPP/17-084
5. Smolen JS, Aletaha D, Barton A, et al. Rheumatoid arthritis. Nat Rev Dis Primers. 2018;4:18001. doi:10.1038/nrdp.2018.1
6. Chang HD, Tokoyoda K, Radbruch A. Immunological memories of the bone marrow. Immunol Rev. 2018;283(1):86–98. doi:10.1111/imr.12656
7. Feuerer M, Beckhove P, Garbi N, et al. Bone marrow as a priming site for T-cell responses to blood-borne antigen. Nat Med. 2003;9(9):1151–1157. doi:10.1038/nm914
8. Nemoto Y, Kanai T, Makita S, et al. Bone marrow retaining colitogenic CD4+ T cells may be a pathogenic reservoir for chronic colitis. Gastroenterology. 2007;132(1):176–189. doi:10.1053/j.gastro.2006.10.035
9. Zou L, Barnett B, Safah H, et al. Bone marrow is a reservoir for CD4+CD25+ regulatory T cells that traffic through CXCL12/CXCR4 signals. Cancer Res. 2004;64(22):8451–8455. doi:10.1158/0008-5472.CAN-04-1987
10. Rudnicka W, Burakowski T, Warnawin E, et al. Functional TLR9 modulates bone marrow B cells from rheumatoid arthritis patients. Eur J Immunol. 2009;39(5):1211–1220. doi:10.1002/eji.200838617
11. Kuca-Warnawin E, Burakowski T, Kurowska W, et al. Elevated number of recently activated T cells in bone marrow of patients with rheumatoid arthritis: a role for interleukin 15? Ann Rheum Dis. 2011;70(1):227–233. doi:10.1136/ard.2009.124966
12. Kuca-Warnawin E, Kurowska W, Prochorec-Sobieszek M, et al. Rheumatoid arthritis bone marrow environment supports Th17 response. Arthritis Res Ther. 2017;19(1):274. doi:10.1186/s13075-017-1483-x
13. Massalska M, Radzikowska A, Kuca-Warnawin E, et al. CD4(+)FOXP3(+) T cells in rheumatoid arthritis bone marrow are partially impaired. Cells. 2020;9:3. doi:10.3390/cells9030549
14. Salomon B, Bluestone JA. Complexities of CD28/B7: CTLA-4 costimulatory pathways in autoimmunity and transplantation. Annu Rev Immunol. 2001;19:225–252. doi:10.1146/annurev.immunol.19.1.225
15. Boasso A, Herbeuval JP, Hardy AW, Winkler C, Shearer GM. Regulation of indoleamine 2,3-dioxygenase and tryptophanyl-tRNA-synthetase by CTLA-4-Fc in human CD4+ T cells. Blood. 2005;105(4):1574–1581. doi:10.1182/blood-2004-06-2089
16. Munn DH, Zhou M, Attwood JT, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281(5380):1191–1193. doi:10.1126/science.281.5380.1191
17. Miwa N, Hayakawa S, Miyazaki S, et al. IDO expression on decidual and peripheral blood dendritic cells and monocytes/macrophages after treatment with CTLA-4 or interferon-gamma increase in normal pregnancy but decrease in spontaneous abortion. Mol Hum Reprod. 2005;11(12):865–870.
18. Murray MF. Tryptophan depletion and HIV infection: a metabolic link to pathogenesis. Lancet Infect Dis. 2003;3(10):644–652. doi:10.1016/S1473-3099(03)00773-4
19. Blair HA, Deeks ED. Abatacept: a Review in Rheumatoid Arthritis. Drugs. 2017;77(11):1221–1233. doi:10.1007/s40265-017-0775-4
20. Bozec A, Luo Y, Engdahl C, Figueiredo C, Bang H, Schett G. Abatacept blocks anti-citrullinated protein antibody and rheumatoid factor mediated cytokine production in human macrophages in IDO-dependent manner. Arthritis Res Ther. 2018;20(1):24. doi:10.1186/s13075-018-1527-x
21. Ngwube A, Shah N, Godder K, Jacobsohn D, Hulbert ML, Shenoy S. Abatacept is effective as GVHD prophylaxis in unrelated donor stem cell transplantation for children with severe sickle cell disease. Blood Adv. 2020;4(16):3894–3899. doi:10.1182/bloodadvances.2020002236
22. Santopaolo M, Sullivan N, Thomas AC, et al. Activation of bone marrow adaptive immunity in type 2 diabetes: rescue by co-stimulation modulator abatacept. Front Immunol. 2021;12:609406. doi:10.3389/fimmu.2021.609406
23. Ciechomska M, Wojtas B, Bonek K, et al. Comprehensive microRNA and transcriptomic profiling of rheumatoid arthritis monocytes: role of microRNA-146b in pro-inflammatory progression. Rheumatology. 2021;60(11):5424–5435. doi:10.1093/rheumatology/keab407
24. Bahraoui E, Serrero M, Planes R. HIV-1 Tat - TLR4/MD2 interaction drives the expression of IDO-1 in monocytes derived dendritic cells through NF-kappaB dependent pathway. Sci Rep. 2020;10(1):8177. doi:10.1038/s41598-020-64847-y
25. Hettinger J, Richards DM, Hansson J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013;14(8):821–830. doi:10.1038/ni.2638
26. Hardin JA. Dendritic cells: potential triggers of autoimmunity and targets for therapy. Ann Rheum Dis. 2005;64(Suppl 4):iv86–90. doi:10.1136/ard.2005.044560
27. Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–324. doi:10.1002/art.1780310302
28. Altman R, Alarcon G, Appelrouth D, et al. The American College of Rheumatology criteria for the classification and reporting of osteoarthritis of the Hip. Arthritis Rheum. 1991;34(5):505–514. doi:10.1002/art.1780340502
29. Kontny E, Szczepanska K, Kowalczewski J, et al. The mechanism of taurine chloramine inhibition of cytokine (interleukin-6, interleukin-8) production by rheumatoid arthritis fibroblast-like synoviocytes. Arthritis Rheum. 2000;43(10):2169–2177.
30. Razmara M, Hilliard B, Ziarani AK, Chen YH, Tykocinski ML. CTLA-4 x Ig converts naive CD4+CD25- T cells into CD4+CD25+ regulatory T cells. Int Immunol. 2008;20(4):471–483.
31. Holmes EW. Determination of serum kynurenine and hepatic tryptophan dioxygenase activity by high-performance liquid chromatography. Anal Biochem. 1988;172(2):518–525. doi:10.1016/0003-2697(88)90478-2
32. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131
33. Kurowska W, Przygodzka M, Jakubaszek M, Kwiatkowska B, Maslinski W. Interleukin-15 as a Biomarker Candidate of Rheumatoid Arthritis Development. J Clin Med. 2020;9(5):1555. doi:10.3390/jcm9051555
34. Terness P, Bauer TM, Rose L, et al. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. J Exp Med. 2002;196(4):447–457. doi:10.1084/jem.20020052
35. Frumento G, Rotondo R, Tonetti M, Damonte G, Benatti U, Ferrara GB. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J Exp Med. 2002;196(4):459–468. doi:10.1084/jem.20020121
36. Nguyen NT, Nakahama T, Le DH, Van Son L, Chu HH, Kishimoto T. Aryl hydrocarbon receptor and kynurenine: recent advances in autoimmune disease research. Front Immunol. 2014;5:551. doi:10.3389/fimmu.2014.00551
37. Merlo LMF, DuHadaway JB, Montgomery JD, et al. Differential Roles of IDO1 and IDO2 in T and B Cell Inflammatory Immune Responses. Front Immunol. 2020;11:1861. doi:10.3389/fimmu.2020.01861
38. Ball HJ, Sanchez-Perez A, Weiser S, et al. Characterization of an indoleamine 2,3-dioxygenase-like protein found in humans and mice. Gene. 2007;396(1):203–213. doi:10.1016/j.gene.2007.04.010
39. Uyttenhove C, Pilotte L, Theate I, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9(10):1269–1274. doi:10.1038/nm934
40. Brandacher G, Perathoner A, Ladurner R, et al. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: effect on tumor-infiltrating T cells. Clin Cancer Res. 2006;12(4):1144–1151. doi:10.1158/1078-0432.CCR-05-1966
41. Mondal A, Smith C, DuHadaway JB, et al. IDO1 is an Integral Mediator of Inflammatory Neovascularization. EBioMedicine. 2016;14:74–82. doi:10.1016/j.ebiom.2016.11.013
42. Gurtner GJ, Newberry RD, Schloemann SR, McDonald KG, Stenson WF. Inhibition of indoleamine 2,3-dioxygenase augments trinitrobenzene sulfonic acid colitis in mice. Gastroenterology. 2003;125(6):1762–1773. doi:10.1053/j.gastro.2003.08.031
43. Sakurai K, Zou JP, Tschetter JR, Ward JM, Shearer GM. Effect of indoleamine 2,3-dioxygenase on induction of experimental autoimmune encephalomyelitis. J Neuroimmunol. 2002;129(1–2):186–196. doi:10.1016/s0165-5728(02)00176-5
44. Szanto S, Koreny T, Mikecz K, Glant TT, Szekanecz Z, Varga J. Inhibition of indoleamine 2,3-dioxygenase-mediated tryptophan catabolism accelerates collagen-induced arthritis in mice. Arthritis Res Ther. 2007;9(3):R50. doi:10.1186/ar2205
45. Davison LM, Liu JC, Huang L, Carroll TM, Mellor AL, Jorgensen TN. Limited effect of Indolamine 2,3-Dioxygenase expression and enzymatic activity on lupus-like disease in B6.Nba2 mice. Front Immunol. 2019;10. doi:10.3389/fimmu.2019.02017
46. Ogbechi J, Clanchy FI, Huang YS, Topping LM, Stone TW, Williams RO. IDO activation, inflammation and musculoskeletal disease. Exp Gerontol. 2020;131:110820. doi:10.1016/j.exger.2019.110820
47. Yasui H, Takai K, Yoshida R, Hayaishi O. Interferon enhances tryptophan metabolism by inducing pulmonary indoleamine 2,3-dioxygenase: its possible occurrence in cancer patients. Proc Natl Acad Sci U S A. 1986;83(17):6622–6626.
48. Liu Y, Liang X, Yin X, et al. Blockade of IDO-kynurenine-AhR metabolic circuitry abrogates IFN-gamma-induced immunologic dormancy of tumor-repopulating cells. Nat Commun. 2017;8:15207. doi:10.1038/ncomms15207
49. Simms PE, Ellis TM. Utility of flow cytometric detection of CD69 expression as a rapid method for determining poly- and oligoclonal lymphocyte activation. Clin Diagn Lab Immunol. 1996;3(3):301–304. doi:10.1128/cdli.3.3.301-304.1996
50. Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J Clin Invest. 2007;117(5):1147–1154. doi:10.1172/JCI31178
51. Wakamatsu E, Omori H, Kawano A, Ogawa S, Abe R. Strong TCR stimulation promotes the stabilization of Foxp3 expression in regulatory T cells induced in vitro through increasing the demethylation of Foxp3 CNS2. Biochem Biophys Res Commun. 2018;503(4):2597–2602. doi:10.1016/j.bbrc.2018.07.021
52. Tarbell KV, Yamazaki S, Steinman RM. The interactions of dendritic cells with antigen-specific, regulatory T cells that suppress autoimmunity. Semin Immunol. 2006;18(2):93–102. doi:10.1016/j.smim.2006.01.009
53. Alvarez-Quiroga C, Abud-Mendoza C, Doniz-Padilla L, et al. CTLA-4-Ig therapy diminishes the frequency but enhances the function of Treg cells in patients with rheumatoid arthritis. J Clin Immunol. 2011;31(4):588–595. doi:10.1007/s10875-011-9527-5
54. Tai X, Cowan M, Feigenbaum L, Singer A. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat Immunol. 2005;6(2):152–162. doi:10.1038/ni1160
55. Schroecksnadel K, Winkler C, Duftner C, Wirleitner B, Schirmer M, Fuchs D. Tryptophan degradation increases with stage in patients with rheumatoid arthritis. Clin Rheumatol. 2006;25(3):334–337. doi:10.1007/s10067-005-0056-6
56. Ozkan Y, Mete G, Sepici-Dincel A, Sepici V, Simsek B. Tryptophan degradation and neopterin levels in treated rheumatoid arthritis patients. Clin Rheumatol. 2012;31(1):29–34. doi:10.1007/s10067-011-1767-5
57. Fallarino F, Grohmann U, Hwang KW, et al. Modulation of tryptophan catabolism by regulatory T cells. Nat Immunol. 2003;4(12):1206–1212. doi:10.1038/ni1003
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.