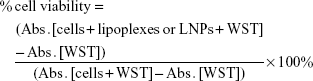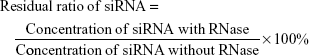Back to Journals » International Journal of Nanomedicine » Volume 12
Effect of the nanoformulation of siRNA-lipid assemblies on their cellular uptake and immune stimulation
Authors Kubota K, Onishi K, Sawaki K, Li T, Mitsuoka K, Sato T, Takeoka S
Received 8 March 2017
Accepted for publication 29 April 2017
Published 19 July 2017 Volume 2017:12 Pages 5121—5133
DOI https://doi.org/10.2147/IJN.S136426
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Kohei Kubota,1,2 Kohei Onishi,3 Kazuaki Sawaki,3 Tianshu Li,4 Kaoru Mitsuoka,5 Takaaki Sato,6 Shinji Takeoka1,3,4
1Cooperative Major in Advanced Biomedical Sciences, Graduate School of Advanced Sciences and Engineering, Waseda University (TWIns), Tokyo, Japan; 2Formulation Research and Phramaceutical Process Group, CMC R&D Center, Kyowa Hakko Kirin Co., Ltd, Shizuoka, Japan; 3Department of Life Science and Medical Bioscience, Graduate School of Advanced Science and Engineering,Waseda University (TWIns), Tokyo, Japan; 4Research Organization for Nano and Life Innovation, Waseda University (TWIns), Tokyo, Japan; 5Research Center for Ultra-High Voltage Electron Microscopy, Osaka University, Osaka, Japan; 6Department of Chemistry and Materials, Faculty of Textile Science and Technology, Shinshu University, Nagano, Japan
Abstract: Two lipid-based nanoformulations have been used to date in clinical studies: lipoplexes and lipid nanoparticles (LNPs). In this study, we prepared small interfering RNA (siRNA)-loaded carriers using lipid components of the same composition to form molecular assemblies of differing structures, and evaluated the impact of structure on cellular uptake and immune stimulation. Lipoplexes are electrostatic complexes formed by mixing preformed cationic lipid liposomes with anionic siRNA in an aqueous environment, whereas LNPs are nanoparticles embedding siRNA prepared by mixing an alcoholic lipid solution with an aqueous siRNA solution in one step. Although the physicochemical properties of lipoplexes and LNPs were similar except for small increases in apparent size of lipoplexes and zeta potential of LNPs, siRNA uptake efficiency of LNPs was significantly higher than that of lipoplexes. Furthermore, in the case of LNPs, both siRNA and lipid were effectively incorporated into cells in a co-assembled state; however, in the case of lipoplexes, the amount of siRNA internalized into cells was small in comparison with lipid. siRNAs in lipoplexes were thought to be more likely to localize on the particle surface and thereby undergo dissociation into the medium. Inflammatory cytokine responses also appeared to differ between lipoplexes and LNPs. For tumor necrosis factor-α, release was mainly caused by siRNA. On the other hand, the release of interleukin-1β was mainly due to the cationic nature of particles. LNPs released lower amounts of tumor necrosis factor-α and interleukin-1β than lipoplexes and were thus considered to be better tolerated with respect to cytokine release. In conclusion, siRNA-loaded nanoformulations effect their cellular uptake and immune stimulation in a manner that depends on the structure of the molecular assembly; therefore, nanoformulations should be optimized before extending studies into the in vivo environment.
Keywords: nanoformulation, siRNA, cryo-TEM, confocal microscopy, endocytosis, immune stimulation
Introduction
In oligonucleotide-based therapeutics, antisense oligonucleotides, small interfering RNAs (siRNA) and aptamers are currently regarded as a new class of agents for remedying hard-to-treat conditions such as cancer, inflammation, and hereditary diseases.1–5 When considering oligonucleotide-based therapeutics, there are several challenges that currently limit the clinical use of siRNA; 1) naked siRNA is unstable in the circulation due to the presence of serum RNase A-type nucleases and due to rapid renal clearance; 2) siRNA can induce immunogenicity, toxicity or off-target gene silencing; and 3) anionic and hydrophilic structures within siRNA prevent its diffusion across cell membranes.6–9 Due to advances in siRNA delivery technology, the development of siRNA-based therapeutics has rapidly progressed with a growing number of current clinical trials.10–14 Lipid-based formulations have been mainly used for systemic administration of siRNA in clinical trials.1–3 Various preparation methods have been proposed; however, the methods that are clinically applied are categorized into 2 types: lipoplexes and lipid nanoparticles (LNPs). Lipoplexes are electrostatic polyelectrolyte complexes formed by mixing preformed cationic liposomes with siRNA in an aqueous environment.15,16 Although lipoplexes have been commonly used, it was reported that siRNA molecules bound to the outer surface of the cationic liposome were prone to dissociate from the liposomal surface and the resulting cationic liposomes aggregated immediately by opsonization in the circulation.17,18 On the other hand, Jeffs et al reported admixture of an aqueous siRNA solution and an alcoholic lipid solution to form siRNA-loaded LNPs in one step with high loading efficiency of siRNA (>80%). The resulting particle size was <200 nm and was monodispersed.19 LNPs have been evaluated in several pipelines of clinical trials as stable nucleic acid lipid particles (SNALP).20,21 Rodrigueza et al reported the preparation of lipid-based nanoparticle (PNT2258) loading of a 24-base DNA oligonucleotide using a one-step method that is similar to the method of SNALP preparation,22 and such nanoparticles have been evaluated in clinical trials.22,23 Though there is no distinct definition of lipoplexes and LNPs, we herein define lipoplexes as electrostatic polyelectrolyte complexes formed by mixing preformed cationic liposomes with siRNA in an aqueous environment, while LNPs are formed by mixing an aqueous siRNA solution with an alcoholic lipid solution in one step. In this study, we prepared two types of siRNA-loaded carrier (lipoplexes and LNPs) with similar particle sizes from components of the same composition using different preparation methods, and evaluated the impact of structure on cellular uptake and immune stimulation.
Materials and methods
Materials
The following lipids were purchased from NOF Co. (Tokyo, Japan): 1,2-dioleyloxy-3-dimethylaminopropane (DODMA), 1,2-distearoyl-sn-glycero-3-phosphatidylethanolamine-N-(polyethylene glycol-2000) (PEG-DSPE), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and cholesterol. Alexa647-labeled PKN3 siRNAs (sense, 5′-AGACUUGAGGACUUCCUGGACAA-3′; antisense, 5′-UUGUCCAGGAAGUCCUCAAGUCU-3′)24 were synthesized by GeneDesign (Osaka, Japan). 3,3′-dihexadecyloxacarbocyanine perchlorate (DiOC16) was purchased from Molecular Probes Inc. (Eugene, OR, USA), whereas chlorpromazine hydrochloride, nystatin and cytochalasin D were purchased from Sigma-Aldrich (St Louis, MO, USA). Alexa488-labeled cholera toxin B was purchased from Molecular Probes Inc. RiboShredder RNase Blend was purchased from Epicentre Biotechnologies (Madison, WI, USA). Proteinase K was purchased from TAKARA BIO INC. (Shiga, Japan). All other reagents used were of analytical grade and were used without further purification.
Preparation of lipoplexes
Lipoplexes were prepared as described previously.15,25 Cationic liposomes were composed of the following lipids: DODMA, PEG-DSPE, DSPC and cholesterol in a molar ratio of 50:1.5:10:38.5. Lipids were dissolved in 3 mL ethanol and then 70.5 μL 1N HCl and 6.93 mL distilled water were added to this mixture (total lipids: 7.8 mg/mL). Cationic liposomes were prepared by extrusion through polycarbonate membrane filters (final pore size: 0.05 μm, LiposoFast, Avestin, Ottawa, ON, Canada) to obtain particles with diameters of ~60 nm. The cationic liposomes were then complexed with Alexa647-labeled siRNA through electrostatic interaction at various N/P ratios (the nitrogen/phosphate [N/P] molar ratio of cationic and anionic charge carriers). Ethanol concentration was reduced to 5% by adding distilled water. Samples were subjected to diafiltration against 300 mL phosphate-buffered saline (PBS) (10 wash volumes) with an Amicon centrifugal ultrafiltration device (MWCO 10,000 Da, Merck Millipore, Tullagreen, Ireland), and sterilized with Acrodisc 0.2-μm syringe filters (Advantec, Tokyo, Japan).
Preparation of LNPs
LNPs were prepared as reported by Jeffs et al.19 An Alexa647-labeled siRNA solution was prepared in 10 mL citrate buffer (20 mM, pH 4.0) to achieve an siRNA concentration of 0.1125 mg/mL. An ethanolic lipid solution was prepared by dissolving DODMA, PEG-DSPE, DSPC, and cholesterol in a molar ratio of 50:1.5:10:38.5 (total lipids: 3.9 mg/mL for N/P ratio of 5), and was added to citrate buffer (20 mM, pH 4.0) to achieve an ethanol concentration of 90 vol%. The siRNA aqueous solution and the lipid aqueous ethanolic solution were mixed together to form particles embedding Alexa647-labeled siRNA. The particles were then immediately diluted with 20 mM citrate buffer containing 300 mM NaCl (pH 6.0). The diluted particles were then incubated at 37°C for 30 min prior to the removal of unencapsulated siRNA by charged membrane filtration using a mustang Q coin filter (Pall Co., Ann Arbor, MI, USA). Ethanol concentration was reduced to 5% by adding distilled water. Samples were concentrated with an Amicon centrifugal ultrafiltration device and subjected to diafiltration against 300 mL PBS. Finally, samples were sterilized with Acrodisc 0.2-μm syringe filters.
Particle size and zeta potential measurements
Size distribution of particles and zeta potential were measured in distilled water with Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). Mean size was determined based on the intensity results. All measurements were performed in triplicate.
Chemical analysis of siRNA and lipid concentrations
The concentrations of Alexa647-labeled siRNA in lipoplexes and LNPs were determined by gradient ion-pair reverse phase high-performance liquid chromatography (HPLC) using an Xbridge OST C18 column (2.1×50 mm, 2.5 μm) set at 60°C. A linear gradient starting at 95% mobile phase A (100 mM hexafluoroisopropanol solution, 8 mM trimethylamine)/5% mobile phase B (methanol) and ending at 70%/30% over 20 min and a flow rate of 0.5 mL/min were used. The amount of Alexa647-labeled siRNA was determined using a peak area count at 260 nm and calculated from an external Alexa647-labeled siRNA standard curve.
Concentrations of the four lipids in lipoplexes and LNPs were determined by gradient reverse-phase HPLC using an Xbridge C8 column (4.6×250 mm, 5 μm) set at 60°C, and 15 min linear gradient from 100% mobile phase A (50% isopropyl alcohol, 20% ammonium acetate [10 mM, pH 7.0], 30% water) to 100% mobile phase B (80% isopropyl alcohol, 20% ammonium acetate [10 mM, pH 7.0]) with a flow rate of 1.0 mL/min. A corona-charged aerosol detector (Corona RS Ultra) (Thermo Scientific, Chelmsford, MA, USA) was used for lipid detection. Individual lipid concentrations were determined from the area count of the elution peak and calculated using an external lipid standard curve with a quadratic fit. Calculation of the N/P ratio was based on the total nitrogen in DODMA versus total phosphate in Alexa647-labeled siRNA from the following equation (1):
|
|
Cryo-transmission electron microscopy (TEM)
Cryo-TEM samples were prepared using 3 μL sample solutions of lipoplexes or LNPs (siRNA: 31 μM) at each N/P ratio. Total lipid concentrations of sample solutions were 17.4 mg/mL at an N/P ratio of 10 and 8.7 mg/mL at an N/P ratio of 5. Samples were added to a standard electron microscope grid with a perforated carbon film. Excess sample solution was removed from the grid by blotting and the grid was then plunge-frozen in liquid ethane to freeze the sample rapidly using a Vitrobot system (FEI, Hillsboro, OR, USA). Images were taken under cryogenic conditions (~88K) at a magnification of 50,000× with an AMT HR CCD camera (Advanced Microscopy Techniques corp., MA, USA). Samples were loaded with a Gatan 70° cryo-transfer holder in an FEI G20 Lab6 200 kV TEM (FEI) under low-dose conditions at an under focus of 4–6 μm to enhance image contrast. Experiments were performed at the Research Center for Ultra-High Voltage Electron Microscopy, Osaka University.
Small-angle X-ray scattering (SAXS)
In the same way as for the cryo-TEM experiments, measurements for lipoplexes and LNPs were performed at an N/P ratio of 5 (siRNA: 31 μM) by a SAXS method.26 All experiments were carried out at Shinshu University using a SAXSess camera (Anton Paar, Austria) in an extended q-range of 0.05–28 nm−1. Samples were filled into a vacuum tight quartz capillary cell. Measured intensities were calibrated for transmission and background scattering was subtracted. A model-independent collimation correction procedure was performed by relying on Lake algorithm.27 Finally, the SAXS profiles in 0.05≤ q/nm−1 ≤8 were further analyzed by the Indirect Fourier Transformation technique.28
Cell culture
HeLa cells (JCRB Cell Bank, Osaka, Japan) or human monocytic cells (THP-1, JCRB Cell Bank) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) or Roswell Park Memorial Institute (RPMI) 1640 Medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich) and 1% penicillin-streptomycin (Gibco). Cells were grown at 37°C in an atmosphere containing 5% CO2.
Cytotoxicity of lipoplexes and LNPs
HeLa cells were seeded in 96-well cell culture plates (5.0×103 cells/well) and incubated in an atmosphere of 5% CO2 at 37°C for 24 h. The medium in the cell culture dish was then exchanged with 100 μL fresh DMEM containing lipoplexes or LNPs (siRNA: 1, 10, 100 and 1,000 nM) in the presence of 10% FBS. After incubation at 37°C for 24 h, cells were washed twice with Dulbecco’s PBS (DPBS, Gibco). Cell viability was tested using a water-soluble tetrazolium salt (WST)-8 assay kit (CCK-8; Dojindo Molecular Technologies, Inc., Osaka, Japan), according to the manufacturer’s instructions. After removal of DPBS, a 100 μL WST-8 solution (CCK-8:DMEM =1:10) was added to each culture followed by a 4 h incubation period. The absorbance of each sample was measured at 450 nm using a microplate reader (Benchmark Plus, Bio-Rad Laboratories Inc, Hercules, CA, USA). Cell viability was calculated from the following equation (2):
|
|
where Abs. and WST represent the absorbance value and WST-8 concentration.
Cellular uptake of lipoplexes and LNPs containing Alexa647-labeled siRNA
HeLa cells were seeded in a 35-mm glass-bottom cell culture dish (1.0×105 cells/dish) and incubated in an atmosphere of 5% CO2 at 37°C for 24 h. Cells were then washed twice with DPBS and 1 mL fresh Hank’s Balanced Salt Solution (HBSS, Gibco) containing lipoplexes or LNPs (siRNA: 100 nM) was added. After incubation at 37°C for various periods (0.5, 1, 2, 6, 12 and 24 h), cells were washed twice with DPBS and fixed with 4% paraformaldehyde in PBS. Cells were observed under a confocal laser scanning microscope (FV1000; Olympus, Tokyo, Japan).29 The overall distribution of siRNA was monitored using Alexa647-labeled siRNA with an excitation wavelength of 635 nm (Alexa647). Average intracellular fluorescence intensity was measured with ImageJ (US National Institutes of Health, MD, USA). Then the fluorescence intensity per unit cell number was evaluated.
HeLa cells (1.0×105 cells) were seeded onto a 24-well plate and incubated in an atmosphere of 5% CO2 at 37°C for 24 h. Cells were then washed twice with DPBS and 1 mL HBSS containing lipoplexes or LNPs (siRNA: 100 nM) was added. After incubation at 37°C for 1 h, cells were harvested and washed with PBS. Then cells were monitored by flow cytometry using a BD LSRFortessa X-20 flow cytometer (BD Biosciences, San Jose, CA, USA).
Intracellular colocalization of siRNA and lipids carriers
To evaluate intracellular colocalization of siRNA and lipid carriers, lipoplexes and LNPs were prepared with an Alexa647-labeled siRNA at an N/P ratio of 5 and the lipophilic dye DiO, to monitor both siRNA and lipid carriers, respectively. DiO was incorporated to a level of 1 mol % of total lipid in both lipoplexes and LNPs. HeLa cells were seeded in a 35 mm glass-bottom cell culture dish (1.0×105 cells/dish) and incubated in an atmosphere of 5% CO2 at 37°C for 24 h. Cells were then washed twice with DPBS and then 1 mL PBS containing lipoplexes or LNPs at an N/P ratio of 5 (siRNA: 100 nM) was added. PBS contained glucose (1.0 mg/mL), CaCl2 (0.14 mg/mL) and MgCl2·6H2O (0.18 mg/mL). After incubation at 37°C for 1 h, cells were washed twice with DPBS and fixed with 4% paraformaldehyde in PBS. Cells were observed in the same way as mentioned above. The overall distribution of DiO was monitored at an excitation wavelength of 473 nm.
RNase protection assay of siRNA in lipoplexes and LNPs
To evaluate RNase protection of siRNA, lipoplexes and LNPs diluted with HBSS were mixed with 25 μL 30 U/mL RiboShredder RNase Blend. Mixtures were then incubated at 37°C for 1 h (siRNA: 1.25 μM). The reaction was terminated by addition of 5 μL proteinase K. siRNA concentrations in lipoplexes and LNPs at an N/P ratio of 5 were determined by the HPLC methods mentioned above. An equivalent amount of free siRNA was used as a control to check RNase activity. The residual ratio of siRNA was calculated from the following equation (3):
|
|
Inhibition studies of endocytosis
HeLa cells were seeded in a 35-mm glass-bottom cell culture dish (1.0×105 cells/dish) and incubated in an atmosphere of 5% CO2 at 37°C for 24 h. Cells were washed with DPBS and then treated with 1 mL DMEM containing inhibitors of endocytosis such as chlorpromazine, nystatin or cytochalasin D (each concentration was 10 μg/mL) for 30 min.30 After cells were washed twice with DPBS, they were treated with 1 mL HBSS containing lipoplexes (siRNA: 1 μM) or LNPs (siRNA: 100 nM) with each inhibitor (10 μg/mL). After incubation at 37°C for 1 h, cells were washed twice with DPBS and fixed with 4% paraformaldehyde in PBS. Cells were observed in the same way as mentioned above. For colocalization studies, HeLa cells (1.0×105 cells) were treated with 1 mL HBSS containing lipoplexes (siRNA: 1 μM) or LNPs (siRNA: 100 nM) together with Alexa488-labeled cholera toxin B (5 μg/mL). After incubation at 37°C for 1 h, cells were processed for microscopy as above.
Immune stimulation
The mature macrophage-like state was induced in THP-1 cells (4.0×105 cells/mL) by treating with a 100 nM phorbol 12-myristate 13-acetate (PMA) solution in dimethyl sulfoxide for 24 h in 12-well plates. PMA was washed off with DPBS and cells were then treated with lipoplexes or LNPs (siRNA: 100 nM). After incubation at 37°C for 24 h, cells were centrifuged at 1,500 rpm for 5 min and the supernatants were collected to evaluate amounts of tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β) released, using a DuoSet enzyme-linked immunosorbent assay (ELISA) kit (R&D systems, Minneapolis, MN, USA). Alternatively, cells were washed twice with DPBS, and TritonX-100 (0.5% in DPBS) was then added to lyse the cells. Intracellular concentrations of lipoplexes and LNPs were determined fluorometrically by measuring siRNA in the lysate with a fluorescence plate reader (excitation and emission wavelengths of 650 and 690 nm, respectively; SpectraMax PARADIGM, Molecular Devices, Japan). The protein concentration of the lysate was determined by a standard protein assay (Pierce 660 nm Protein Assay: Thermo Fisher Scientific, Waltham, MA, USA).30 Cellular uptake efficiencies of lipoplexes and LNPs were evaluated as ng Alexa647-labeled siRNA per μg protein.
Statistical analysis
The student’s t-test was applied in the analysis of RNase protection assay. The Aspin–Welch test was applied in the comparison of cellular uptake of Alexa647-labeled siRNA evaluated by flow cytometry. Inflammatory cytokine production was analyzed by one-way analysis of variance followed by Dunnett’s multiple-comparisons test. Statistically significant differences were identified when P<0.05.
Results
Preparation and characterization of lipoplexes and LNPs
Particle size, polydispersity and zeta potential were measured using Malvern Zetasizer. As shown in Table 1, particle size and zeta potential changed depending on the N/P ratio. Cationic liposomes used for lipoplexes were prepared by an extrusion method using polycarbonate membrane filters (final pore size: 0.05 μm) before complexation with siRNA. The resulting cationic liposomes were 60 nm in diameter and exhibited a zeta potential of ~40 mV. As previously reported,15,25 size was found to increase slightly on complexation with siRNA molecules and zeta potentials also decreased accordingly. Particle size showed a maximum at N/P ratio of 5. On the other hand, the zeta potential increased from 10 to 29 mV as N/P ratio increased from 2 to 10. The above trends were similar to those reported elsewhere.31,32 While LNPs were prepared in one step, siRNA and lipid solutions were mixed together. LNP sizes were 90–120 nm smaller than those of lipoplexes and were similar to those reported using the same method.19,33 As with lipoplexes, LNP size showed a maximum at an N/P ratio of 5 and then decreased at an N/P ratio of 10. The zeta potential increased from 12 to 33 mV as the N/P ratio increased from 2 to 10. LNPs showed a similar trend to lipoplexes, though they showed higher potential values than lipoplexes at each N/P ratio.
Cryo-TEM
Lipoplexes observed on cryo-TEM appeared to be aggregated and deformed in the aggregate (Figure S1A and C). They seemed larger than LNPs, corresponding to the particle size results obtained with Zetasizer. On the other hand, LNPs appeared to adopt more compact and solid structures, which were dispersed, thereby maintaining an isolated spherical structure, rather than an aggregated form (Figure S1B and D).
Small-angle X-ray scattering
The diffraction maxima q =1 nm−1 for both lipoplexes and LNPs at an N/P ratio of 5 indicated a lamellar organization with a periodicity d of ~6.0 nm (d =2π/q) (Figure S2A). The average diameters of lipoplexes and LNPs at an N/P ratio of 5 were calculated to be ~80 and 65 nm, respectively (Figure S2B).
Cytotoxicity of lipoplexes and LNPs
Cytotoxicity against HeLa cells was evaluated for lipoplexes and LNPs at an N/P ratio of 5. The rate of viability was assessed using a WST reduction assay.34 No cytotoxicity was evident over the concentration range of siRNA used (1–1,000 nM), corresponding to a DODMA concentration range of 0.24–240 nM (Figure S3).
Cellular uptake of lipoplexes and LNPs containing Alexa647-labeled siRNA
To analyze cellular uptake of siRNA-loaded carriers, a red-fluorescent (Alexa647)-labeled siRNA was monitored by fluorescence microscopy. HeLa cells were treated with lipoplexes or LNPs containing 100 nM Alexa647-labeled siRNA at an N/P ratio of 5, for up to 24 h. As shown in Figure 1, the amount of siRNA delivered by LNPs was significantly greater than that by lipoplexes. The Alexa647-labeled siRNA delivered by LNPs was detected intracellularly 30 min after adding sample to the cells and the fluorescence intensity per unit cell number showed a maximum in 12 h. Same as LNPs, the fluorescence intensity of siRNA delivered by lipoplexes was saturated after 12 h, but the values achieved were about 4 times lower than those obtained with LNPs. It showed the same trend when evaluated by flow cytometry. Mean fluorescent intensity of LNPs showed much higher than that of lipoplexes at an N/P ratio of 5 and a dose of 100 nM siRNA for 1 h (Figure S4). In both LNPs and lipoplexes, cellular uptake efficiency increased when the N/P ratio was raised from 2 to 10 (Figure S5). However, the average intracellular fluorescence intensity of siRNA delivered by LNPs was much higher than that by lipoplexes at each N/P ratio. In comparison, free Alexa647-labeled siRNA incubated with HeLa cells for 1 h, exhibited no fluorescence, either within cells or in the extracellular medium, suggesting that siRNA was not internalized and was removed by washing (Figure S6).
Intracellular colocalization of siRNA and lipids carriers
Particle size, polydispersity and zeta potential were not significantly changed after incorporation of DiO into both lipoplexes and LNPs at an N/P ratio of 5 (Table S1). HeLa cells were treated with lipoplexs or LNPs for 1 h (siRNA: 100 nM). Fluorescence microscopy results indicated that intracellular dynamics of siRNA and DiO differed completely between lipoplexes and LNPs (Figure 2). Both siRNA and DiO were efficiently taken up by cells in the case of LNPs, and the intracellular colocalization of these compounds was confirmed. On the other hand, intracellular intensity of the DiO signal in lipoplexes was weaker than that in LNPs; that of siRNA in lipoplexes was much weaker. These data correspond to the results shown in Figure 1, indicating that the uptake of siRNA was very weak 1 h after the addition of lipoplexes to the cells.
RNase protection assay of siRNA in lipoplexes and LNPs
We first confirmed that naked siRNA was completely degraded by RNase during a 1 h incubation at 37°C after the addition of RNase. We then evaluated the residual ratios of siRNA in lipoplexes and LNPs under the same conditions. As shown in Figure 3, almost 100% of the siRNA remained in LNPs, while ~80% remained in lipoplexes. It is suggested that siRNA in the LNPs was completely protected against RNase while siRNA in the lipoplexes was partially degraded.
Inhibition studies of endocytosis
To study the mechanism of endocytosis, HeLa cells were pretreated with chlorpromazine, an inhibitor of clathrin-mediated endocytosis (CME), nystatin, an inhibitor of caveolae-mediated endocytosis (CvME), or cytochalasin D, an inhibitor of macropinocytosis, as indicated in methods. Because the cellular uptake efficiency of lipoplexes was much lower than that of LNPs (as shown in the above cellular uptake experiments), HeLa cells were treated with lipoplexes at an siRNA dose of 1 μM, which is 10 times higher than the dose used in the case of LNPs. Chlorpromazine did not inhibit the internalization of either lipoplexes or LNPs (Figure S7). In the case of lipoplexes, cellular uptake of siRNA was weakly reduced on treatment with nystatin or cytochalasin D, suggesting that internalization was driven by both CvME and macropinocytosis. On the other hand, in the case of LNPs, cellular uptake of siRNA was strongly reduced on treatment with nystatin, indicating that cellular uptake with the LNPs was predominantly driven by CvME. In colocalization studies, we used Alexa488-labeled cholera toxin as a marker of CvME. In the case of LNPs, the intracellular colocalization of Alexa488-labeled cholera toxin with Alexa647-labeled siRNA suggested that internalization of LNPs was driven by CvME (Figure S8). On the other hand, in the lipoplexes case, the intracellular localization of siRNA and cholera toxin was not completely merged, indicating that lipoplexes were partially taken up by CvME. These observations agreed with the evaluation using inhibitors. Interestingly, it was confirmed that in both lipoplexes and LNPs, the addition of nystatin inhibited localization of siRNA near cell surface regions, while the addition of cytochalasin D inhibited the localization of siRNA near central regions of the cell; thus, different uptake pathways influenced intracellular behaviors of siRNA (Figure S7).
Immune stimulation
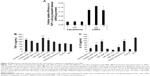
It has been reported that cellular uptake of siRNA-loaded nanoformulation induced activation of the innate immune response.6,9 To evaluate the inflammatory cytokine response mediated by lipoplexes or LNPs, cellular uptake efficiency by THP-1 cells and amounts of TNF-α and IL-1β released from cells were measured. Similar to the cellular uptake behavior of HeLa cells, LNPs internalized siRNA molecules into THP-1 cells at significantly high efficiencies when compared with lipoplexes (Figure 4A). The extents of TNF-α release triggered by lipoplexes or LNPs were higher than those obtained with cationic liposomes that did not carry siRNA (Figure 4B). Furthermore, the amounts released by both lipoplexes and LNPs increased with decreases in N/P ratio, indicating that the release of TNF-α was mainly caused by siRNA; this was because the amount of siRNA loaded per particle became larger as the N/P ratio decreased. The amounts of IL-1β release caused by lipoplexes or LNPs are shown in Figure 4C. Cationic liposomes caused a greater release of IL-1β than the negative control (PBS), to the same level as the adenosine triphosphate positive control. Moreover, when the N/P ratios of lipoplexes increased, the amount of IL-1β increased almost to the same level as with the cationic liposomes, indicating that the release of IL-1β was mainly caused by the cationic properties of particles.
Discussion
In this study, we prepared 2 types of siRNA-loaded carriers: lipoplexes and LNPs, with similar particle sizes, using lipid components of the same composition. Since these carriers were thought to form molecular assemblies of differing structures, the impact of structure on their cellular uptake and immune stimulation was evaluated. Lipoplexes were formed by adding siRNA to preformed cationic liposomes 60 nm in diameter; the resulting lipoplexes were found to be 130–150 nm in diameter, which was slightly larger than that of LNPs (100–120 nm). These results were corroborated by SAXS measurements: again, diameters were similar, though size estimates were smaller, namely 80 nm for lipoplexes and 60 nm for LNPs. SAXS measurements also indicated that both lipoplexes and LNPs at an N/P ratio of 5 exhibited lamellar organizations inside the particles, suggesting that both particles adopted similar structures even though their diameters differed. In addition, the number of correlated bilayers in lipoplexes seemed higher to some extent in view of the sharper interference peak located at ca. 1 nm−1. Thus, it is suggested that the structure of lipoplexes cannot be described simply in terms of attachment of siRNA to the liposomal surface, but is considered to adopt a lamellar structure as reported elsewhere.35–38 Aggregation of deformed particles was observed in lipoplexes by examination of cryo-TEM data, while LNP particles exhibited stable spherical structures. These differences may be explained by differences in zeta potentials. In fact, measured N/P values of LNPs were slightly higher than those of lipoplexes at loading N/P ratios of 5 and 10 (Table 1). However, even at the loading N/P ratio of 2, where the measured N/P value in lipoplexes was higher than that in LNPs, the zeta potential of LNPs was higher than that of lipoplexes. Considering the above discussion, it is suggested that the siRNA molecules of lipoplexes were more localized on the particle surface than those of LNPs, resulting in the lower zeta potential and aggregation tendency.
The efficiencies of siRNA uptake by HeLa cells or THP-1 cells differed between lipoplexes and LNPs. We found that cellular uptake of LNPs incorporating siRNA was significantly higher than that of lipoplexes. As described above, the measured N/P value was higher in LNPs than in lipoplexes at an N/P ratio of 5; therefore, the greater numbers of positive LNPs were considered to reflect higher cell internalization.32,39 However, as shown in Figure S5, regardless of N/P ratios and zeta potentials, LNPs showed markedly high intracellular uptake in comparison with lipoplexes, which is thought to be due to the structural differences between particles. We hypothesized that the cellular uptake mechanism would therefore be different. In this experiment, we stained siRNA and lipid carriers separately and evaluated their intracellular dynamics. In the case of LNPs, both Alexa647-labeled siRNA and DiO were taken up 1 h after addition of LNPs to the cells and showed colocalization, indicating that both were incorporated together into the cells in a co-assembled state. However, only small amounts of DiO and siRNA uptake were observed in the case of lipoplexes. Under conditions of comparable DiO fluorescence intensity, it was clear that lipoplexes delivered smaller amounts of siRNA to cells than LNPs (Figure 2B), suggesting that siRNA in lipoplexes was partially dissociated from the particles. Compared with siRNA in LNPs, siRNA in lipoplexes is thought to be more likely to localize on the particle surface; therefore, siRNA tended to dissociate under the influence of divalent cations dissolved in the medium.19,31
In support of the points discussed above, RNase was used to investigate the localization of siRNA within the 2 nano formulations. The greater degradation of siRNA in lipoplexes by RNase suggested that a proportion of siRNA bound to the surface of the lipoplexes (as reported elsewhere18) would be dissociated and digested by RNase. Therefore, the results herein indicated that the lipoplexes were more susceptible to the external environment of the particles, and were thus considered to be less favorable as a carrier of siRNA than the LNPs from the viewpoint of administration for in vivo evaluation.
Since a variety of uptake pathways into cells are known for siRNA-loaded carriers and since intracellular behavior should depend on such pathways, we evaluated the uptake mechanism in lipoplexes and LNPs using endocytosis inhibitors. Non-viral gene vectors, including siRNA-loaded carriers cannot easily pass through the cell membrane because of their size or hydrophilic nature, and most are taken up by an endocytotic route.40–42 Endocytosis is roughly divided into pinocytosis, so-called cell drinking, and phagocytosis, so-called cell eating.43 Phagocytosis is an actin-based uptake mechanism mainly handled by phagocytes, such as macrophages.44 In contrast, pinocytosis is a common uptake mechanism for all cells that can be further divided into 4 possible processes: CME, CvME, macropinocytosis and other clathrin- and caveolae-independent endocytoses.43–45 Endocytosis is known to be greatly affected by the size of the particles: CME, 100–120 nm; CvME, 50–100 nm; macropinocytosis, ~5 μm; and clathrin- and caveolae-independent endocytosis, 40–50 nm.42–44 The mechanisms whereby lipoplexes and LNPs are internalized are not well understood and seem to be affected by their physicochemical characteristics and the nature of the target cells.42,44,46 Specific inhibitors for each pathway are also known: chlorpromazine for CME, nystatin for CvME, and cytochalasin D for macropinocytosis.30,42 Two groups have reported internalization, trafficking and endosomal escape of siRNA delivered by LNPs composed of different lipids.47 Sahay et al reported that LNPs composed of synthetic lipidoid C12-200 were internalized into HeLa cells by macropinocytosis when monitored with Alexa647-labeled siRNA.48 Gilleron et al studied LNPs with siRNA that was labeled by either fluorescent dyes or gold nanoparticles and evaluated their trafficking in different cells.49 LNPs were found to enter the cells through both macropinocytosis and CME.49 For the lipoplexes and LNPs used in our study, cellular uptake of LNPs was inhibited by nystatin, indicating that CvME was the major route. On the other hand, cellular uptake of lipoplexes was inhibited by nystatin and cytochalasin D, indicating both CvME and macropinocytosis as the major routes. Cellular uptake of the cationic liposomes used to prepare lipoplexes was inhibited by nystatin and cytochalasin D (data not shown); thus, the cellular uptake mechanisms of lipoplexes, where a part of siRNA would be dissociated from the particle surface in particular media conditions, is suggested to be related to the cationic liposomes.
In this study, the use of inhibitors produced certain interesting cell internalization behaviors. In lipoplexes, when nystatin was added, siRNA molecules were distributed in the central regions of cells and not distributed near the peripheral regions of the cell membrane. Distribution in the former manner would be due to macropinocytosis, while the latter manner would occur via CvME that was inhibited by nystatin. When cytochalasin D was added, siRNA distribution showed a ring-like structure namely, the central regions were dark, suggesting that siRNA molecules were internalized by CvME and macropinocytosis was inhibited. This is because caveolae are highly stable,42,46 and CvME is considered to be a very slow process in comparison with macropinocytosis.42,50 In summary, lipoplexes would not be internalized effectively by CvME and the contribution of macropinocytosis would be relatively increased. On the other hand, LNPs were actively internalized by CvME. We consider that cholesterol, which is a known requirement for CvME,51,52 locates near the surface of LNPs and can be recognized preferentially by CvME receptors; however, further detailed analyses are needed to confirm this. From this study, the nanoformulations obtained by different preparation methods influence mechanisms of cellular uptake or changes to intracellular dynamics, which would also affect efficacy profiles. As with cellular uptake efficiency, intracellular dynamics of siRNA and lipids, especially endosome escape efficiency, would be factors that greatly affect the onset of efficacy. We are currently studying endosome escape and cytosol delivery of siRNA after cell internalization of LNPs.
Regarding inflammatory cytokine responses against lipoplexes and LNPs, the amounts of TNF-α and IL-1β released showed different profiles. Judge et al reported that lipid-complexed siRNA induced inflammatory cytokines through the activation of toll like receptors (TLRs) 3, 7 and 8, whereas naked siRNA did not.53 This indicates that the inflammatory response is related to cell internalization of siRNA-loaded carriers because these TLRs are known to localize on the endosomal membrane.6,9,53 On the other hand, Kedmi et al reported cytokine release, including both TNF-α and IL-1β by positively charged lipid nanoparticles via stimulation of TLR4 in addition to immune stimulation by nucleic acids, and TLR4 is known to localize on the cell surface.54 In this study, because the released amount of TNF-α was higher as the N/P ratio was lower, TLRs 3, 7, and 8 in the cells that recognize the nucleic acid were likely to be involved. However, the involvement of TLR4 could not be denied. In contrast, siRNA does not appear to be responsible for the release of IL-1β. Stimulated release of IL-1β by lipoplexes is postulated to be a result of the cationic nature of particles because of which siRNA is dissociated from the lipid carriers when incorporated into THP-1 cells. Therefore, it is thought that the cationic nature of liposomes stimulates TLR4 and increases the amount of IL-1β released, even if the uptake efficiency of lipoplexes into the cells is low. Therefore, it is necessary to consider toxicity of siRNA-loaded nanoformulations, including immune stimulation by both siRNA and the cationic nature of particles and also to consider preparation methods.
Conclusion
Most of studies performed focused on the types of cationic lipid used and the composition of the preparation affect intracellular uptake or toxicity. In this study, we prepared 2 types of nanoformulations (lipoplexes and LNPs) with similar particle sizes from the same components with the same composition using different preparation methods, and evaluated the impact of their structures on cellular uptake and immune stimulation. LNPs showed higher cellular uptake efficiency of siRNA than lipoplexes; however, there were no significant differences in physicochemical properties when dispersed in PBS after preparation using lipid components of the same composition. Stability of the lipoplex–siRNA interaction was affected by external environments such as the medium, whereas LNPs were less susceptible to the external environment. Furthermore, LNPs were also better tolerated in terms of reduced cytokine (TNF-α and IL-1β) release in response to siRNA and cationic lipid carriers. LNPs released lower amounts of TNF-α and IL-1β than lipoplexes. Thus, LNPs are considered to be more beneficial siRNA carriers following their intravenous administration.
Disclosure
The authors report no conflicts of interest in this work.
References
Zuckerman JE, Davis ME. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat Rev Drug Discov. 2015;14(12):843–856. | ||
Wu SY, Lopez-Berestein G, Calin GA, Sood AK. RNAi therapies: drugging the undruggable. Sci Transl Med. 2014;6(240):240ps7. | ||
Wicki A, Witzigmann D, Balasubramanian V, Huwyler J. Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J Control Release. 2015;200:138–157. | ||
Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2(12):711–719. | ||
Wittrup A, Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet. 2015;16(9):543–552. | ||
Kanasty RL, Whitehead KA, Vegas AJ, Anderson DG. Action and reaction: the biological response to siRNA and its delivery vehicles. Mol Ther. 2012;20(3):513–524. | ||
Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33(9):941–951. | ||
Zhang Y, Satterlee A, Huang L. In vivo gene delivery by nonviral vectors: overcoming hurdles? Mol Ther. 2012;20(7):1298–1304. | ||
Buyens K, De Smedt SC, Braeckmans K, et al. Liposome based systems for systemic siRNA delivery: stability in blood sets the requirements for optimal carrier design. J Control Release. 2012;158(3):362–370. | ||
Ozcan G, Ozpolat B, Coleman RL, Sood AK, Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv Drug Deliv Rev. 2015;87:108–119. | ||
Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J. 2011;6(9):1130–1146. | ||
Wang Y, Miao L, Satterlee A, Huang L. Delivery of oligonucleotides with lipid nanoparticles. Adv Drug Deliv Rev. 2015;87:68–80. | ||
Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11(1):59–67. | ||
Caster JM, Patel AN, Zhang T, Wang A. Investigational nanomedicines in 2016: a review of nanotherapeutics currently undergoing clinical trials. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2017;9(1):1–18. | ||
Santel A, Aleku M, Keil O, et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13(16):1222–1234. | ||
Wong FM, Reimer DL, Bally MB. Cationic lipid binding to DNA: characterization of complex formation. Biochemistry. 1996;35(18):5756–5763. | ||
Meyer O, Kirpotin D, Hong K, et al. Cationic liposomes coated with polyethylene glycol as carriers for oligonucleotides. J Biol Chem. 1998;273(25):15621–15627. | ||
Buyens K, Lucas B, Raemdonck K, et al. A fast and sensitive method for measuring the integrity of siRNA-carrier complexes in full human serum. J Control Release. 2008;126(1):67–76. | ||
Jeffs LB, Palmer LR, Ambegia EG, Giesbrecht C, Ewanick S, MacLachlan I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm Res. 2005;22(3):362–372. | ||
Coelho T, Adams D, Silva A, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369(9):819–829. | ||
Tabernero J, Shapiro GI, LoRusso PM, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3(4):406–417. | ||
Rodrigueza WV, Woolliscroft MJ, Ebrahim AS, et al. Development and antitumor activity of a BCL-2 targeted single-stranded DNA oligonucleotide. Cancer Chemother Pharmacol. 2014;74(1):151–166. | ||
Tolcher AW, Rodrigueza WV, Rasco DW, et al. A phase 1 study of the BCL2-targeted deoxyribonucleic acid inhibitor (DNAi) PNT2258 in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2014;73(2):363–371. | ||
Aleku M, Schulz P, Keil O, et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008;68(23):9788–9798. | ||
Maurer N, Wong KF, Stark H, et al. Spontaneous entrapment of polynucleotides upon electrostatic interaction with ethanol-destabilized cationic liposomes. Biophys J. 2001;80(5):2310–2326. | ||
Sato T, Sakai H, Sou K, Medebach M, Glatter O, Tsuchida E. Static structures and dynamics of hemoglobin vesicle (HBV) developed as a transfusion alternative. J Phys Chem B. 2009;113(24):8418–8428. | ||
Glatter OJ. A new method for the evaluation of small-angle scattering data. J Appl Crystallogr. 1977;10(5):415–421. | ||
Lake JA. An iterative method of slit-correcting small angle x-ray data. Acta Cryst. 1967;23:191–194. | ||
Li T, Takeoka S. A novel application of maleimide for advanced drug delivery: in vitro and in vivo evaluation of maleimide-modified pH-sensitive liposomes. Int J Nanomedicine. 2013;8:3855–3866. | ||
Li T, Takeoka S. Enhanced cellular uptake of maleimide-modified liposomes via thiol-mediated transport. Int J Nanomedicine. 2014;9:2849–2861. | ||
Xu Y, Hui SW, Frederik P, Szoka FC. Physicochemical characterization and purification of cationic lipoplexes. Biophys J. 1999;77(1):341–353. | ||
Zhang Y, Li H, Sun J, et al. DC-Chol/DOPE cationic liposomes: a comparative study of the influence factors on plasmid pDNA and siRNA gene delivery. Int J Pharm. 2010;390(2):198–207. | ||
Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release. 2005;107(2):276–287. | ||
Pérez SE, Gándola Y, Carlucci AM, González L, Turyn D, Bregni C. Formulation strategies, characterization, and in vitro evaluation of lecithin-based nanoparticles for siRNA delivery. J Drug Deliv. 2012;2012:986265–986274. | ||
Huebner S, Battersby BJ, Grimm R, Cevc G. Lipid-DNA complex formation: reorganization and rupture of lipid vesicles in the presence of DNA as observed by cryoelectron microscopy. Biophys J. 1999;76(6):3158–3166. | ||
Weisman S, Hirsch-Lerner D, Barenholz Y, Talmon Y. Nanostructure of cationic lipid-oligonucleotide complexes. Biophys J. 2004;87(1):609–614. | ||
Kapoor M, Burgess DJ, Patil SD. Physicochemical characterization techniques for lipid based delivery systems for siRNA. Int J Pharm. 2012;427(1):35–57. | ||
Dan N, Danino D. Structure and kinetics of lipid-nucleic acid complexes. Adv Colloid Interface Sci. 2014;205:230–239. | ||
Chono S, Li SD, Conwell CC, Huang L. An efficient and low immunostimulatory nanoparticle formulation for systemic siRNA delivery to the tumor. J Control Release. 2008;131(1):64–69. | ||
Friend DS, Papahadjopoulos D, Debs RJ. Endocytosis and intracellular processing accompanying transfection mediated by cationic liposomes. Biochim Biophys Acta. 1996;1278(1):41–50. | ||
Zuhorn IS, Kalicharan R, Hoekstra D. Lipoplex-mediated transfection of mammalian cells occurs through the cholesterol-dependent clathrin-mediated pathway of endocytosis. J Biol Chem. 2002;277(20):18021–18028. | ||
Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58(1):32–45. | ||
Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422(6927):37–44. | ||
Hillaireau H, Couvreur P. Nanocarriers’ entry into the cell: relevance to drug delivery. Cell Mol Life Sci. 2009;66(17):2873–2896. | ||
Lamaze C, Schmid SL. The emergence of clathrin-independent pinocytic pathways. Curr Opin Cell Biol. 1995;7(4):573–580. | ||
Medina-Kauwe LK, Xie J, Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene Ther. 2005;12(24):1734–1751. | ||
Wang Y, Huang L. A window onto siRNA delivery. Nat Biotechnol. 2013;31(7):611–612. | ||
Sahay G, Querbes W, Alabi C, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31(7):653–658. | ||
Gilleron J, Querbes W, Zeigerer A, et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31(7):638–646. | ||
Mundy DI, Machleidt T, Ying YS, Anderson RG, Bloom GS. Dual control of caveolar membrane traffic by microtubules and the actin cytoskeleton. J Cell Sci. 2002;115(Pt 22):4327–4339. | ||
Martin S, Parton RG. Caveolin, cholesterol, and lipid bodies. Semin Cell Dev Biol. 2005;16(2):163–174. | ||
Bae YU, Huh JW, Kim BK, Park HY, Seu YB, Doh KO. Enhancement of liposome mediated gene transfer by adding cholesterol and cholesterol modulating drugs. Biochim Biophys Acta. 2016;1858(12):3017–3023. | ||
Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23(4):457–462. | ||
Kedmi R, Ben-Arie N, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;31(26):6867–6875. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.