Back to Journals » Cancer Management and Research » Volume 12
Effect of LINC00657 on Apoptosis of Breast Cancer Cells by Regulating miR-590-3p
Authors Shan Q, Qu F, Yang W, Chen N
Received 13 February 2020
Accepted for publication 21 April 2020
Published 15 June 2020 Volume 2020:12 Pages 4561—4571
DOI https://doi.org/10.2147/CMAR.S249576
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Qiuli Shan,1 Fan Qu,1 Weiping Yang,2 Ningning Chen1
1College of Biological Science and Technology, University of Jinan, Jinan 250022, People’s Republic of China; 2Department of Thyroid Breast Surgery, Qingdao Chengyang People’s Hospital, Qingdao, People’s Republic of China
Correspondence: Qiuli Shan
College of Biological Science and Technology, University of Jinan, No. 336, Nanxinzhuan West Road, Jinan, Shandong Province 250022, People’s Republic of China
Tel +86-18753151252
Email [email protected]
Objective: To investigate the effect of LINC00657 on breast carcinoma by regulating miR-590-3p.
Methods: Ninety-seven cases with breast carcinoma who were admitted to Qingdao Chengyang People’s Hospital were collected. The breast carcinoma (n=97) and tumor-adjacent tissues (n=97) of patients were collected during the operation with the permission of the patients. The expressions of LINC00657 and miR-590-3p were detected in breast carcinoma cells and tissues. The breast carcinoma cells were transfected and their proliferation, migration, invasion and apoptosis were detected.
Results: LINC00657 was highly expressed in breast carcinoma tissues, while miR-590-3p was reduced (P< 0.05). The proliferation, invasion and migration of cells transfected with si-LINC00657 or miR-590-3p-mimics were significantly inhibited, and the apoptosis rate increased, resulting in the up-regulation of the expressions of apoptosis-related proteins Bax and Caspase-3 and the reduction of Bcl-2 (P< 0.05). After si-LINC00657 or miR-590-3p-mimics, the level of GOLPH3 decreased. Through double luciferase report and RIP experiment, it was confirmed that LINC00657 could act as a sponge of miR-590-3p to negatively regulate its expression. After correlation analysis, it was concluded that there was a negative correlation between LINC00657 and miR-590-3p. Rescue experiments concluded that co-transfection of si-LINC00657+miR-590-3P-inhibitor could reverse the inhibitory action of si-LINC00657 on breast carcinoma cells.
Conclusion: LINC00657 can participate in the biological behavior process of breast carcinoma by regulating miR-590-3p/GOLPH3 signal.
Keywords: LINC00657, miR-590-3p, GOLPH3, breast carcinoma, apoptosis
Introduction
In recent years, breast carcinoma has become the most common disease among women worldwide, with a high incidence rate. It is considered to be the primary factor of cancer-related death among women.1,2 According to statistics, 250,000 new cases and 40,000 deaths are estimated in the USA in 2017.3
According to the latest epidemiological statistics, the incidence of breast carcinoma exceeded 2 million and the death toll was approximately 600,000 worldwide in 2018.4 Globally, breast carcinoma has become one of the malignant tumors that seriously threaten human life and health due to the rapid raise in the incidence of breast carcinoma in the past decades.5 At present, the main treatment methods for breast carcinoma include surgery, chemotherapy, radiotherapy, immunotherapy, etc., which can improve the prognosis of patients to a certain extent, but there are still some limitations.6,7 At present, the main obstacles in the treatment of breast carcinoma may be caused by over-activation of oncogenic signaling pathways, inhibition of tumor suppressor genes expression or dormancy, and subsequent reawakening of residual tumor cells, resulting in decreased sensitivity to treatment.8 Therefore, we urgently need to properly regulate the expression of tumor suppressor genes or carcinogenic genes to solve the related problems in the traditional treatment of breast carcinoma.
Long non-coding RNA (lncRNA) is defined as a kind of non-coding RNA with a length greater than 200bp. In recent years, whether lncRNA can be used as a regulator of various biological processes has become the focus of attention.9 More and more studies have indicated that lncRNA participates in the biological behavior process of various diseases, and it is tight related to the development and progression of diseases.10 LINC00657, located in cytoplasm, is an lncRNA with a length of 5343 nucleotides, highly conserved in mammalian body sequence and low coding ability. It is also called DNA damage activated non-coding RNA (NORAD).11 Some related studies have suggested that LINC00657 expression is abnormal in breast carcinoma, and the up-regulation of LINC00657 is related to the overall poor prognosis of breast carcinoma.12 However, whether LINC00657 can participate in the regulation of breast carcinoma biological process still needs more research evidence. At present, many studies have demonstrated that lncRNA can act as a molecular sponge for miR (microRNA), thus participating in the regulation of various biological processes.13 miR-590-3p plays an anti-carcinoma in various cancers.14 Studies by RohiniM15 and others have indicated that over-expression of miR-590-3p can inhibit proliferation and increase apoptosis of breast carcinoma cells. Moreover, miR-590-3p can be involved in the occurrence and development of breast cancer by regulating a variety of protein pathways. Golgi phosphoprotein 3 (GOLPH3) is a novel biomarker associated with poor prognosis of breast cancer and resistance to chemotherapy, and it has been reported to be involved in the biological processes of a variety of diseases due to its strong tumorigenicity.16 Moreover, the latest research has suggested that miR-590-3p can immediately target the 3’-UTR of GOLPH3 mRNA to inhibit its expression, thus regulating the malignant behavior of breast carcinoma cells.17 It is suggested that miR-590-3p/GOLPH3 can be used as an effective pathway to mediate the progression of breast cancer.
At present, there are few studies on whether LINC00657 can participate in the regulation of miR-590-3p/GOLPH3 pathway. So in this paper, the expression of LINC00657 in breast carcinoma was detected to observe the impact of LINC00657 on the biological behavior of breast carcinoma cells and its regulatory mechanism, so as to provide an effective target and reference for the clinical treatment of breast cancer.
Methods
Tissues Collection
Ninety-seven cases with breast carcinoma who underwent surgical treatment in Qingdao Chengyang People’s Hospital were selected. With the consent of the patients, 97 cases of carcinomas and 97 cases of adjacent tissues were collected for follow-up study. The experiment has been authorized by the ethics committee of Qingdao Chengyang People’s Hospital, and the subjects and their families were described in detail. The subjects agreed and signed a full informed consent.
Inclusion criteria: Clinical data were complete and patients could actively cooperate with the work; Patients with breast carcinoma were diagnosed by imaging and pathology; No other malignant tumors in patients occurred at the time of admission.
Exclusion criteria: Patients were not accompanied by their families at the time of admission and had autoimmune system defects. The patient had mental illness and refused to provide laboratory samples.
Cells Culture and Transfection
Breast carcinoma cell lines MCF-7, MDA-MB-231, T47D, BT-549 and normal breast epithelial cell line MCF-10A were all from the ATCC cell bank in the United States without mycoplasma contamination by detection. The cell line was put into DMEM medium comprising 10% fetal bovine serum (GIBCO) and cultured at 37°C and 5% CO2. When the adherent growth and fusion of cells were observed to reach 85%, 25% pancreatin was added for digestion. After digestion, the cell line was placed into the medium for continuous culture and passage. Then, the expressions of LINC00657 and miR-590-3p in each cell line were detected. MCF-7 and MDA-MB-231 at logarithmic growth phase were collected for transfection. The miR-590-3p-inhibitor (inhibition sequence), miR-590-3p-mimics (over-expression sequence), miR-negative control (miR-NC), targeted inhibition of LINC00657 (si-LINC00657), blank control (Vector) were transfected, respectively. The Lipofectamine™ 2000 kit (Invitrogen) was used to transfect the cells. The operating procedure was strictly in accordance with the kit instructions.
Detection of qRT-PCR
First, total RNA was obtained from tissues and cells. The procedure was performed according to the instructions of TRIzol kit (Invitrogen, USA). The purity, concentration and integrity of the extracted total RNA were tested by UV spectrophotometer and agarose gel electrophoresis. The reverse transcription was carried out by TaqMan™ Reverse transcription kit (Invitrogen, USA). The transcription steps were strictly operated according to the kit instructions. The obtained cDNA was further studied. 7500 PCR (ABI) and SYBR Premix Ex Taq kit (TaKaRa) were used. PRC amplification cycle conditions were as follows: pre-degeneration at 95°C for 60s, degeneration at 95°C for 30s, anneal and extension at 60°C for 40s, then followed a total of 40 cycles. In the experiment, three parallel repeating holes were designed, and all specimens were repeatedly tested for 3 times. LINC00657 and GOLPH3 used GAPDH as internal reference, and miR-590-3p used U6 as internal reference. The specific sequence is shown in Table 1. The results were calculated by 2-ΔΔct.
 |
Table 1 Primers Sequence |
Detection of Western Blot
Cells were obtained and proteins were lysed by RIPA buffer (Cell Signal Technology, Inc., MA, USA). The concentration and quantification of protein were determined by BCA kit (Beyotime Biotechnology, Shanghai, China). Electrophoretic separation was performed with 10%SDS gel, and protein was transferred to PVDF (EMD Millipore Corporation, Billerica, MA, USA), and allowed to stand at ambient temperature for 1 h (closed with 5% skim milk-PBS solution). The first antibody was added (Santa Cruz Biotechnology, USA) and cultivated overnight at 4°C. The first antibody was removed by washing the film, and HRP-conjugated goat anti-rabbit second antibody (Abcam, USA) was added, incubated at 37°C for 1h, and rinsed 3 times with PBS for 5 min each time. Finally, the blot was detected with ECL chemiluminescence reagent (Thermo Fisher Scientific, USA) and the Amersham Prime ECL Plus detection system was used.
Detection of Cell Proliferation Ability
The proliferation activity was evaluated by CCK-8 assay kit (Dojindo), and the processed cells were inoculated into 96-hole plates at a density of 1000 cells/hole. After cultivation for 24–72 hours, CCK-8 reagent was put into cells and cultivated at ambient temperature for 2 hours. Then, optical density values were surveyed at 450nm to ascertain cell proliferation capability.
Detection of Cell Invasion and Migration Ability
Transwell technology (Corning, USA) was used for detection. Cells were adjusted to 5*105 and inoculated into the 24-hole Transwell chamber at the top. Then, 600 μL complete medium was put into lower chamber. After cultivation for 24h, the migrated cells were fixed, then dyed with 0.1% of crystal violet and numbered under confocal microscope. For invasive assays, invasive cells were tested with Transwell Matrigel (BD Biosciences, USA) invasive assays. Matrigel (BD Biosciences, USA) was precoated into the upper chamber. Other test steps were semblable to migration determination.
Detection of Apoptosis Ability
Apoptosis was tested by Annexin V-FITC /PI apoptosis detection kit (Thermo Fisher Scientific). The different processed cells were washed with cold phosphate saline buffer and digested with trypsin. Then, cells were re-suspended in a 100μL binding buffer (comprising FITC-Annexin V/PI) and hatched in darkness for 15min. FACScan flow cytometer (Becton Dickinson Company, USA) was applied to analyze the apoptosis. This test was repeated for 3 times to take the average.
Determination of Double Luciferase
StarBase 3.0 was used to predict the target gene. The pmirGLO-LINC00657-wt and pmirGLO-LINC00657-mut vectors were constructed and co-transfected into cells with miR-590-3p and NC, respectively. After transfection for 48 h, luciferase intensity was detected by double luciferase reporter genes (Solarbio, CA, China). The operating procedure was strictly in accordance with the instructions.
RNA-Binding Protein Co-Immunoprecipitation (RIP Experiment)
Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, USA) was applied for detection. The RIP lysis buffer was used to lyse the cells. Then, 100 μL whole cell extract was cultivated overnight at 4°C with magnetic beads bound to human anti-Ago2 antibody or normal mouse IgG (Millipore, USA). The protein in the sample was digested with proteinase K to extract immunoprecipitation RNA. Finally, the purified RNA was analyzed by qRT-PCR to verify the existence of binding targets.
RNA Fluorescence in situ Hybridization (FISH)
FISH analysis was performed using Ribo ™ fluorescence in situ hybridization kit (Ribobio Company, China). The counting probe was purchased from Empire Genomics and labeled with fluorescent dye. RNA FISH was performed using a fluorescence in situ hybridization kit in accordance with the manufacturer’s instructions. Fluorescence detection was carried out by AxioObserver Z1 microscope (Zeiss).
Statistical Processing
In this study, SPSS20.0 was applied for statistical analysis of the collected data. GraphPad 7 was used to draw the required pictures. The t-test was applied to compare the differences between the two groups. One-way ANOVA was used for comparison among groups. LSD-t test was used for pairwise comparison afterwards. The association between LINC00657 and miR-590-3p was analyzed by Pearson test. There was statistical difference with p<0.05.
Results
Expressions of LINC00657 and miR-590-3p in Breast Carcinoma
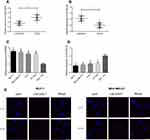
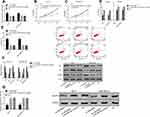
PCR detection results indicated that compared with the adjacent tissue of breast cancer, the LINC00657 expression in breast carcinoma tissue was up-regulated, while miR-590-3p was down-regulated (shown in Figure 1) (P<0.001). Further detection demonstrated that the LINC00657 expression was up-regulated in breast carcinoma cell lines (P<0.05), while the miR-590-3p expression was low (P<0.05). In addition, we conducted an RNA-FISH study with a probe, and the results showed that LINC00657 was mainly located in the cytoplasm of breast cancer cells.
Effect of LINC00657 on Biological Behavior of Breast Carcinoma Cells
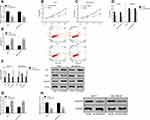
Breast carcinoma cell lines were transfected, and the LINC00657 expression in the cell line transfected with si-LINC00657 reduced significantly (P<0.05). The biological behavior of transfected cells was detected (Figure 2). It could be derived that the proliferation, invasion and migration of MCF-7 and MDA-MB-231 cells transfected with si-LINC00657 were inhibited, and the apoptosis rate increased, resulting in the enhancement of the expressions of apoptosis-related proteins Bax and Caspase-3 and the decrease of Bcl-2 expression (P<0.05). Moreover, silencing the LINC00657 expression could lead to an increase in miR-590-3p level and a decrease in GOLPH3 protein level (P<0.05).
Impact of miR-590-3P on Biological Behavior of Breast Carcinoma Cells
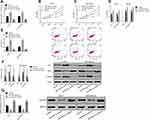
The breast carcinoma cell lines were transfected, the cell line miR-590-3p expression was enhanced after miR-590-3p-mimics transfection, and the cell line miR-590-3p expression was down-regulated after miR-590-3p- inhibitor transfection (P<0.05). The biological behavior of transfected cells was detected (Figure 3). It could be found that the proliferation, invasion and migration ability of the cells transfected by miR-590-3p-mimics were inhibited, and the apoptosis rate was increased, leading to the enhancement of the expression of apoptosis-related proteins Bax and Caspase-3, the decrease of Bcl-2 expression (P<0.05), and the decrease of GOLPH3 protein level (P<0.05). However, the result was opposite after transfection by miR-590-3p- inhibitor.
Targeted Relationship Between LINC00657 and miR-590-3p
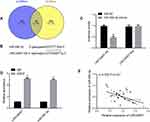
In order to understand the relevant mechanism of LINC00657, we predicted the target genes of LINC00657 through starbase and IncBase, and found 28 target genes. According to the previous research results and the current literature and data, there was a targeted binding site between LINC00657 and miR-590-3p. Furthermore, through the double luciferase report detection, LINC00657-wt and LINC00657-mut vectors were constructed and co-transfected into cells with miR-590-3p and NC, respectively. It was found that miR-590-3p could inhibit the fluorescence activity of LINC00657-Wt (P<0.05). Moreover, RIP experiment indicated that compared with anti-IgG, the enrichment content of LINC00657 and miR-590-3p was significantly enhanced after immunoprecipitation with anti-Ago2 antibody. The correlation between the expression of LINC00657 and miR-590-3p in 97 breast cancer tissues was analyzed. Then, Pearson test showed that there was a negative correlation between the LINC00657 and miR-590-3p expression in breast carcinoma (r=−0.525, P<0.001). (Figure 4)
Rescue Experiment
In order to further verify that LINC00657 could affect the biological function of breast carcinoma cells by regulating miR-590-3p, si-LINC00657 and miR-590-3p- inhibitor were co-transfected into breast carcinoma cell lines MCF-7 and MDA-MB-231 (Figure 5). Compared with NC, there was no obvious difference in cell proliferation, invasion, migration, apoptosis and related proteins after co-transfection of si-LINC00657+miR-590-3p-inhibitor (P>0.05). But compared with si-LINC00657 group, the cell proliferation, invasion and migration ability increased significantly and the apoptosis rate decreased significantly after co-transfection, while the expression of apoptosis-related proteins Bax and Caspase-3 decreased, and Bcl-2 expression enhanced (P<0.05). It was concluded that si-LINC00657 reduced the expression of GOLPH3 protein, while co-transfection of si-LINC00657+miR-590-3p-inhibitor reversed the phenomenon.
Discussion
The research on lncRNA in breast carcinoma is increasing, which provides an important new direction for molecular biology and clinical medicine research. LncRNA has potential as a novel molecular marker and has broad clinical application prospects.18 “Sponge” theory holds that one of the bases for lncRNA to play a role in tumors is to form ceRNA network with miRNA.19 lncRNA can directly participate in the regulation of target genes. Because of its long length, the core seed sequence contained in LNC RNA can be adsorbed to the corresponding miRNA, which affects the abundance of target gene mRNA, thus affecting gene expression.20
After quantitative detection by qRT-PCR, the results of this study demonstrated that LINC00657 was enhanced in breast carcinoma tissues and cells, suggesting that LINC00657 plays a carcinogenic role in breast carcinoma and may participate in the occurrence of breast carcinoma. Moreover, since the cell biological function and the potential molecular action on lncRNA were closely related to its localization, the localization of LINC00657 was detected by RNA-FISH experiment. It was found that LINC00657 was mainly located in the cytoplasm of cells, which was similar to previous studies.21 To further explore the role of LINC00657, cell experiments were further studied. The expression of LINC00657 had been silenced, and it was found that silencing the expression of LINC00657 could significantly hinder the proliferation, invasion and migration of breast carcinoma cells, leading to an increase in the apoptosis rate, an enhancement of the apoptosis-related proteins Bax and Caspase-3 expressions, and a decrease in the expression of Bcl-2 (P<0.05). Previous studies have found that LINC00657 is highly expressed in colorectal cancer,22 glioblastoma23 and non-small cell lung cancer,24 which is consistent with this study. Moreover, studies by Sun25 and others have indicated that knocking out LINC00657 can inhibit migration and proliferation of esophageal cancer cells and increase radiation sensitivity. LINC00657 can act as competitive endogenous RNA (ceRNA) to increase JunB expression by binding to miR-615-3p and participate in biological regulation of tumor cells. Therefore, combined with the results of this study, LINC00657 can be regarded as a potential therapy target for breast carcinoma. However, the further mechanism of LINC01087 is still unclear.
At present, many literatures have suggested that lncRNA can be used as ceRNA to regulate RNA transcripts through competition with miR (microRNA) and promote changes in biological functions of various tumor cells.26,27 There are reports that lncRNA XIST can regulate the expression of CDX1 by mediating miR-155, so as to affect the progress of breast carcinoma.28 Studies by Li29 et al have indicated that lncRNA GAS5 can inhibit the proliferation and invasion of triple negative breast carcinoma cells by competitively binding miR-196A-5P. In this study, we conducted on-line predictive analysis of potential targeted miR of LINC00657 and found that there were binding sites between LINC00657 and miR-590-3p. miRNAs become a critical part of gene silencing and can play a role in gene regulation networks.30 Research indicated that miR-590-3p was disordered in various cancers, and it can inhibit the development and progression of bladder cancer31 and hepatocellular carcinoma32 and plays a role of oncogene in prostate cancer.33 In this research, miR-590-3p expression was low in breast carcinoma, which is consistent with the results of Abdolvahabi34 and others, indicating that miR-590-3p has an anti-carcinoma effect in breast carcinoma. Subsequently, we over-expressed and inhibited miR-590-3p, and it was concluded that miR-590-3p obviously inhibited the proliferation and metastasis of breast carcinoma cells after over-expression. The apoptosis rate of tumor cells was increased by up-regulating Bax, Caspase-3 expression and down-regulating Bcl-2 expression (P<0.05). The results demonstrated that miR-590-3p can regulate the generation and progression of breast carcinoma cells. Double luciferase report and RIP experiment confirmed that LINC00657 could act as a sponge of miR-590-3p to negatively regulate its expression. Through correlation analysis, it was found that there was a negative correlation of LINC00657 with miR-590-3p, which was laterally confirmed that LINC00657 could regulate miR-590-3p.
Previous researches have proved that miR-590-3p can directly target GOLPH3, and over-expression of GOLPH3 prevented the inhibition of miR-590-3p proliferation. The results revealed that miR-590-3p/GOLPH3 signaling pathway could regulate the proliferation of breast carcinoma cell.17 Therefore, we speculated that LINC00657 might regulate the development of breast carcinoma by mediating miR-590-3p/GOLPH3 signaling pathway. Most literatures indicated that GOLPH3 was a novel oncogene, and its content expression was closely related to tumor proliferation and metastasis.35 High expression of GOLPH indicated poor prognosis of epithelial ovarian cancer.35 Previous studies revealed that over-expression of GOLPH3 promoted proliferation of breast cells.36 In this research, by detecting the expression of GOLPH3 after different transfection, it was found that silencing LINC00657 or over-expression of miR-590-3p would lead to a decline of GOLPH3 level. Through rescue experiments, it was found that silencing LINC00657 expression could promote the down-regulation of GOLPH3, while co-transfection of miR-590-3p inhibitor reversed the effect. Combined with the results of this experimental study, it was revealed that LINC00657 could participate in the regulation of malignant behaviors such as proliferation, invasion, migration and apoptosis of breast carcinoma cells through miR-590-3p/GOLPH3 axis.
This research was mainly designed to discuss the mechanism of LINC00657 in breast carcinoma, but there are still some limitations. LINC00657 has many regulatory networks in tumors, and whether it is possible to mediate the development and progression of breast carcinoma through another way, and whether LINC00657 has inhibitory effect on the growth of tumorigenesis test in nude mice needs further experiments to explore.
To sum up, LINC00657 can participate in the biological behavior process of breast carcinoma by regulating miR-590-3p.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (NSFC) [grant number 21507040], the Natural Science Foundation of Shandong Province [grant number BS2015SW023].
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Robbins AS, Lin CC, et al. Factors that contributed to black-white disparities in survival among nonelderly women with breast cancer between 2004 and 2013. J Clin Oncol. 2018;36(1):14–24. doi:10.1200/JCO.2017.73.7932
2. DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. doi:10.3322/caac.21412
3. Busch EL, Whitsel EA, Kroenke CH, Yang YC. Social relationships, inflammation markers, and breast cancer incidence in the women’s health initiative. Breast. 2018;39:63–69. doi:10.1016/j.breast.2018.03.013
4. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
5. Spronk I, Schellevis FG, Burgers JS, de Bock GH, Korevaar JC. Incidence of isolated local breast cancer recurrence and contralateral breast cancer: a systematic review. Breast. 2018;39:70–79. doi:10.1016/j.breast.2018.03.011
6. Song M, Vogelstein B, Giovannucci EL, Willett WC, Tomasetti C. Cancer prevention: molecular and epidemiologic consensus. Science. 2018;361(6409):1317–1318. doi:10.1126/science.aau3830
7. Aguirre-Ghiso JA. How dormant cancer persists and reawakens. Science. 2018;361(6409):1314–1315. doi:10.1126/science.aav0191
8. Pang B, Wang Q, Ning S, et al. Landscape of tumor suppressor long noncoding RNAs in breast cancer. J Exp Clin Cancer Res. 2019;38(1):79. doi:10.1186/s13046-019-1096-0
9. Filippov-Levy N, Cohen-Schussheim H, Trope CG, et al. Expression and clinical role of long non-coding RNA in high-grade serous carcinoma. Gynecol Oncol. 2018;148(3):559–566. doi:10.1016/j.ygyno.2018.01.004
10. Mahmoudian-Sani MR, Jalali A, Jamshidi M, et al. Long non-coding RNAs in thyroid cancer: implications for pathogenesis, diagnosis, and therapy. Oncol Res Treat. 2019;42(3):136–142. doi:10.1159/000495151
11. Lee S, Kopp F, Chang TC, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164(1–2):69–80. doi:10.1016/j.cell.2015.12.017
12. Liu H, Li J, Koirala P, et al. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget. 2016;7(15):20584–20596. doi:10.18632/oncotarget.7828
13. Arunkumar G, Deva Magendhra Rao AK, Manikandan M, et al. Expression profiling of long non-coding RNA identifies linc-RoR as a prognostic biomarker in oral cancer. Tumour Biol. 2017;39(4):1010428317698366. doi:10.1177/1010428317698366
14. Tong H, Zhuang X, Cai J, et al. Long noncoding RNA ZFAS1 promotes progression of papillary thyroid carcinoma by sponging miR-590-3p and upregulating HMGA2 expression. Onco Targets Ther. 2019;12:7501–7512. doi:10.2147/OTT.S209138
15. Rohini M, Gokulnath M, Miranda PJ, Selvamurugan N. miR-590-3p inhibits proliferation and promotes apoptosis by targeting activating transcription factor 3 in human breast cancer cells. Biochimie. 2018;154:10–18. doi:10.1016/j.biochi.2018.07.023
16. Tang S, Pan H, Wei W, Yang H, Liu J, Yang R. GOLPH3: a novel biomarker that correlates with poor survival and resistance to chemotherapy in breast cancer. Oncotarget. 2017;8(62):105155–105169. doi:10.18632/oncotarget.21927
17. Song Q, Chen Q, Wang Q, et al. ATF-3/miR-590/GOLPH3 signaling pathway regulates proliferation of breast cancer. BMC Cancer. 2018;18(1):255. doi:10.1186/s12885-018-4031-4
18. Liu Y, Guo G, Zhong Z, et al. Long non-coding RNA FLVCR1-AS1 sponges miR-155 to promote the tumorigenesis of gastric cancer by targeting c-Myc. Am J Transl Res. 2019;11(2):793–805.
19. Cao K, Fang Y, Wang H, Jiang Z, Guo L, Hu Y. The lncRNA HOXA11-AS regulates Rab3D expression by sponging miR-125a-5p promoting metastasis of osteosarcoma. Cancer Manag Res. 2019;11:4505–4518. doi:10.2147/CMAR.S196025
20. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
21. Elguindy MM, Kopp F, Goodarzi M, et al. PUMILIO, but not RBMX, binding is required for regulation of genomic stability by noncoding RNA NORAD. Elife. 2019;8. doi:10.7554/eLife.48625.
22. Shaker OG, Ali MA, Ahmed TI, et al. Association between LINC00657 and miR-106a serum expression levels and susceptibility to colorectal cancer, adenomatous polyposis, and ulcerative colitis in Egyptian population. IUBMB Life. 2019;71(9):1322–1335. doi:10.1002/iub.2039
23. Chu L, Yu L, Liu J, et al. Long intergenic non-coding LINC00657 regulates tumorigenesis of glioblastoma by acting as a molecular sponge of miR-190a-3p. Aging (Albany NY). 2019;11(5):1456–1470. doi:10.18632/aging.101845
24. Zhang R, Niu Z, Pei H, Peng Z. Long noncoding RNA LINC00657 induced by SP1 contributes to the non-small cell lung cancer progression through targeting miR-26b-5p/COMMD8 axis. J Cell Physiol. 2019. doi:10.1002/jcp.29222
25. Sun Y, Wang J, Pan S, et al. LINC00657 played oncogenic roles in esophageal squamous cell carcinoma by targeting miR-615-3p and JunB. Biomed Pharmacother. 2018;108:316–324. doi:10.1016/j.biopha.2018.09.003
26. Pilyugin M, Irminger-Finger I. Long non-coding RNA and microRNAs might act in regulating the expression of BARD1 mRNAs. Int J Biochem Cell Biol. 2014;54:356–367. doi:10.1016/j.biocel.2014.06.018
27. Fan H, Lv Z, Gan L, et al. A novel lncRNA regulates the toll-like receptor signaling pathway and related immune function by stabilizing FOS mRNA as a competitive endogenous RNA. Front Immunol. 2019;10:838. doi:10.3389/fimmu.2019.00838
28. Zheng R, Lin S, Guan L, et al. Long non-coding RNA XIST inhibited breast cancer cell growth, migration, and invasion via miR-155/CDX1 axis. Biochem Biophys Res Commun. 2018;498(4):1002–1008. doi:10.1016/j.bbrc.2018.03.104
29. Li S, Zhou J, Wang Z, Wang P, Gao X, Wang Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed Pharmacother. 2018;104:451–457. doi:10.1016/j.biopha.2018.05.056
30. Sun ZQ, Shi K, Zhou QB, et al. MiR-590-3p promotes proliferation and metastasis of colorectal cancer via Hippo pathway. Oncotarget. 2017;8(35):58061–58071. doi:10.18632/oncotarget.19487
31. Mo M, Peng F, Wang L, Peng L, Lan G, Yu S. Roles of mitochondrial transcription factor A and microRNA-590-3p in the development of bladder cancer. Oncol Lett. 2013;6(2):617–623. doi:10.3892/ol.2013.1419
32. Ge X, Gong L. MiR-590-3p suppresses hepatocellular carcinoma growth by targeting TEAD1. Tumour Biol. 2017;39(3):1010428317695947. doi:10.1177/1010428317695947
33. Chen H, Luo Q, Li H. MicroRNA-590-3p promotes cell proliferation and invasion by targeting inositol polyphosphate 4-phosphatase type II in human prostate cancer cells. Tumour Biol. 2017;39(3):1010428317695941. doi:10.1177/1010428317695941
34. Abdolvahabi Z, Nourbakhsh M, Hosseinkhani S, et al. MicroRNA-590-3P suppresses cell survival and triggers breast cancer cell apoptosis via targeting sirtuin-1 and deacetylation of p53. J Cell Biochem. 2019;120(6):9356–9368. doi:10.1002/jcb.28211
35. Sun J, Yang X, Zhang R, et al. GOLPH3 induces epithelial-mesenchymal transition via Wnt/beta-catenin signaling pathway in epithelial ovarian cancer. Cancer Med. 2017;6(4):834–844. doi:10.1002/cam4.1040
36. Tenorio MJ, Ross BH, Luchsinger C, et al. Distinct biochemical pools of golgi phosphoprotein 3 in the human breast cancer cell lines MCF7 and MDA-MB-231. PLoS One. 2016;11(4):e0154719. doi:10.1371/journal.pone.0154719
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.