Back to Journals » OncoTargets and Therapy » Volume 9
Effect of evodiamine and berberine on miR-429 as an oncogene in human colorectal cancer
Authors Liu H, Huang C, Wu L, Wen B
Received 21 January 2016
Accepted for publication 31 March 2016
Published 6 July 2016 Volume 2016:9 Pages 4121—4127
DOI https://doi.org/10.2147/OTT.S104729
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Hong Liu, Chao Huang, Liyun Wu, Bin Wen
Institute of Spleen and Stomach, Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, People’s Republic of China
Abstract: Loss of epithelial organization and deregulated microRNAs are hallmarks of malignant carcinomas, but the relationship between them has been poorly understood. This study was designed to investigate the changes in the expression of E-cadherin, Par3, and miR-429 during the development of human colorectal cancer (CRC). E-cadherin and Par3 levels were quantitatively detected by immunohistochemistry and Western blotting. An in vitro culture of colorectal tissue was established to analyze the effect of berberine (BER) and evodiamine (EVO) on the level of miR-429. Our results suggested that E-cadherin and Par3 were remarkably decreased in tumor tissues compared with those in normal tissues, and miR-429 was upregulated in tumor tissues. After treatment of BER and EVO, the level of miR-429 was lower in tumor tissues than in normal tissues. This study investigated the potential relationship between miR-429, E-cadherin, and Par3 in CRC. The data suggested that BER and EVO can be potential therapeutic agents for CRC, as they downregulated the expression level of miR-429.
Keywords: microRNAs, tissue culture
Introduction
Colorectal cancer (CRC) with a multistep process is a leading cause of high cancer-related morbidity and mortality, with >600,000 deaths worldwide annually.1 Although various etiologies have been proposed, the aberrant accumulation of genetic and epigenetic changes is an important mechanism during tumorigenesis. Numerous data show that altered epigenetic modification contributes to carcinogenesis.2 MicroRNAs are small noncoding RNAs that can lead to the translational repression of target genes. The increasing interest in the microRNAs with regulatory function has led to continued research; hence, many new microRNAs have been found as oncogenes or tumor suppressors with high sensitivity and specificity.3 MicroRNAs take part in a large number of biological activities, including cell differentiation, proliferation, and apoptosis.4,5 Altered microRNAs are found in various cancers, including breast cancer,6 lung cancer,7 gastric cancer,8 hepatocellular carcinoma,9 pancreatic cancer,10 and esophageal squamous cell cancer.11 Some studies have discovered many aberrant microRNAs in CRC,12 which seem to show the significant roles of microRNAs in the development of CRC. The expression and relative functions of E-cadherin and Par3, which might be regulated by miR-429, are still unclear.
Berberine (BER) is widely used in clinical practice in antidiarrheal, antihypertensive, and anti-inflammatory diseases.13–16 Furthermore, increasing evidence supports that BER can suppress chemical-induced carcinogenesis, tumor development, and invasion.16–18 Evodiamine (EVO) has been reported to exert the antitumor, antinociceptive, and vasorelaxant effect.19,20 EVO can suppress tumor invasion and promote apoptosis.21 However, the antitumor mechanisms of BER and EVO are still poorly understood.
Our study aimed to discover the potential biomarkers for CRC and investigate the role of potential biomarkers and microRNA in tumorigenesis.
Materials and methods
Subjects
Tissues resected operationally were from the First Affiliated Hospital of Guangdong University of Traditional Chinese Medicine (TCM). Patients had not accepted any radiotherapy or chemotherapy before operation. The diagnosis of CRC was confirmed by at least two pathologists. The collected samples were immediately put into liquid nitrogen for conservation. Tumor tissues and matched tumor-adjacent normal tissues were collected from the same patient. The study was approved by the Institutional Ethics Committee of Guangzhou University of TCM and performed in accordance with the ethical guidelines of the Declaration of Helsinki. Informed written consent was obtained from all participants.
MicroRNA target prediction
MicroRNA target genes were predicted by retrieving databases of miRBase and miRanda, and targets can be found at (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/, http://www.microrna.org/microrna/home.do, http://mirdb.org/miRDB/). The prediction was made to find potential target genes with a high degree of sequence matching and stable secondary structure.
Western blotting
Tissues were collected, lysed, and blotted as described previously.22 Membranes were blocked with blocking solution (5% skim milk in Tris-buffered saline with Tween 20) and incubated with primary antibody E-cadherin (Abcam, Cambridge, MA, USA) and Par-3 (BIOSS, Beijing, China), followed by incubation with appropriate secondary antibody. The densitometry of Western blotting results was measured using the ImageJ software (National Institutes of Health, Bethesda, USA).
Immunohistochemistry
Specimens were confirmed by hematoxylin and eosin-stained sections. Formalin-fixed, paraffin-embedded sections (4 μm) were deparaffinized in xylene, rehydrated in graded alcohol, and rinsed in phosphate-buffered saline (PBS). Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol for 20 minutes. Epitope retrieval was performed in citrate buffer for 5 minutes at 100°C. Slides were incubated with mouse monoclonal primary antibodies (BIOSS) diluted to 1:30 at 4°C overnight. After washing three times with fresh PBS, the sections were subsequently incubated with secondary antibody at room temperature (37°C) for 30 minutes. For visualization of the reaction, the diaminobenzidine-tetrahydrochloride was stained, then counterstained with hematoxylin, dehydrated and cover slipped. Two observers without knowledge of the studies’ data performed evaluation of the staining.
Tissue culture and treatment
After washing by Hanks, the tissues were overlaid with advanced Dulbecco’s Modified Eagle’s Medium/F12 containing 100 U/mL penicillin (Thermo Fisher Scientific, Waltham, MA, USA), 100 μ/mL streptomycin (Thermo Fisher Scientific). BER (Sigma-Aldrich Co, St Louis, MO, USA) and EVO (WakoPure Chemical Industries, Ltd., Osaka, Japan) were added to the basal medium, respectively, after 48 hours. Tissues were treated with BER at 4, 8, 16 μM, EVO at 3, 6, 12 μM for 72 hours. EVO was dissolved in dimethyl sulfoxide. The concentration of dimethyl sulfoxide was <0.1% in the medium.
Cultured tissue staining
Calcein-AM (Dojindo Laboratories, Kumamoto, Japan) and propidium iodide (PI) (Dojindo) solution were mixed and prepared with PBS. A portion of 1 mL Calcein-AM/PI solution was supplemented onto the culture medium. The obtained tissues were incubated at 37°C for 20 minutes and then the tissue samples were washed twice with the PBS. The tissue slices were observed under the fluorescence microscope (Olympus, Tokyo, Japan).
Quantitative reverse transcription polymerase chain reaction
Random primer 10 μmol/L was added to total RNA 1 μg, then diethylpyrocarbonate was blended for 10 minutes, finally the reverse transcription reaction liquids (5× RT reaction buffer 5 μL, deoxynucleotide 1 mmol/L, RNase inhibitor 1 U/μL, M-MLV RTase 8 U/μL (Invitrogen, Waltham, MA, USA), diethylpyrocarbonate 4 μL) were added for 1 hour to obtain reverse transcription products. For quantitative reverse transcription polymerase chain reaction (qRT-PCR), SYBR Green I (Ribobio, Guangzhou, People’s Republic of China) was used in accordance with instructions, relative expression levels were calculated by subtracting the average U6 (internal control) Ct values and applying the formula 2−ΔΔCt. All samples were performed in triplicate. The used primers by Primer 5.0 ABI PRISM7900 system are listed in Table 1. 2× qRT-PCR reaction mix 8 μL (2× master mix 5 μL, PCR sequence specific primer F 0.5 μL, PCR sequence specific primer R 0.5 μL) were added to holes corresponding to 384-PCR plate, then corresponding 2 μL complementary DNA was added. Three-step program was used for the PCR reaction, and after the reaction, analysis of melting curves was performed.
Statistical analysis
All data analysis was performed by Stata 12.0 software (StataCorp LP, College Station, TX, USA). The qRT-PCR results are presented as the mean ± standard error of the mean. One-way analysis of variance analysis was adapted to accomplish the comparison among the three groups, assuming normal distribution; if not, nonparametric test was performed. Significant level α was 0.05 and P-value <0.05 was considered to be statistically significant.
Results
MicroRNAs target prediction and functional analysis
Predictive information from three databases, miRBase, miRanda, and Targets can be performed; overlapping parts of the three predictive results were served as final result predicted. Furthermore, relative functional analysis of target genes was made as well. Consequently, a large number of target genes mediated by miR-429 and relative functions were acquired. As illustrated in Figure 1, a large quantity of target genes was mediated by miR-429.
  | Figure 1 A network of target miRNAs–messenger RNAs. Blue nodes represent target genes and yellow nodes represent miRNAs. |
Hematoxylin and eosin staining
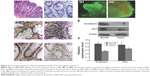
One of the earliest histopathological phenomena of CRC is aberrant crypt foci (ACF). Increasing evidence supported that ACF is the early indication of colon cancer.23 The size and number of ACF contributes to CRC development. Normal colon crypts were cut vertically to observe their histopathological characteristics. The colon crypts without branching or protuberances were test tube shaped and smooth on the surface. Compared to normal tissues, the crypts tumor tissues were losing their morphology (Figure 2A).
Immunohistochemistry and Western blotting
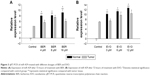
Immunohistochemical staining suggested that E-cadherin and Par-3 were expressed in all tissue specimens, and were dominantly observed in the cytoplasm and membrane (Figure 2B and C). The tissues were cultured for 3 and 25 days and subsequently stained with Calcein-AM/PI (Figure 2D). Western blotting showed that the expression levels of E-cadherin and Par-3 in the tumor tissues were significantly lower than those in the normal tissues (Figure 2E and F). MicroRNAs in the tissues treated with BER and EVO were also detected by qRT-PCR. The expression levels of miR-429 were significantly upregulated in all the treatment groups compared with those in the control group. However, the level of miR-429 in tumor tissues was considerably lower than that in normal tissues treated with BER and EVO, suggesting that the expression level of miR-429 was regulated by BER and EVO treatment, as illustrated in Figure 3.
Discussion
Inheritance mediated by epigenetics plays a crucial role in carcinogenesis. Some studies have discovered that the degree of epigenetic difference between cancer and normal cells was considerably higher than that between normal cells with different phenotypes and even different germ cells, such as fibroblasts and epithelial cells, suggesting that epigenetics exerts a pivotal effect on tumorigenesis. In addition, complexity and frequency of epigenetic alterations observed in cancer cells appear to be able to overturn the explanation that epigenetic alterations rely on a single event;24 on the contrary, these pathological alterations may be caused by multiple mechanisms. Previous studies have shown that aberrant microRNAs changes play a vital role in different steps of cancer formation.25 Difference of microRNA profiling in the different phases of colorectal canceration reveals that oncogenesis is not a molecular event but a sequential process involving a number of genes and signaling molecules. Furthermore, two or more microRNAs may participate in the canceration process at the same time. Some studies reported that microRNAs expression levels are globally reduced in cancer patients,26 many researchers have found the opposite in CRC specimens. Our previous study found upregulated miR-200a-5p, miR-429, miR-497-5p, and downregulated miR-185-5p, miR-625-3p, miR-1237 in tumor tissues. These results reveal that tumors significantly differ from normal tissues in epigenetics, which is consistent with the previous studies. Among the six microRNAs, expression level of miR-429 was significantly increased in tumor tissues compared with normal tissues. Higher expression levels of miR-429 correlate with a poor prognosis in patients with advanced-stage ovarian carcinoma.27 MiR-429 may regulate many proteins, including zinc finger E-box binding homeobox 1), DNA methyltransferase, fibroblast growth factor and RNA binding motif protein, etc. Overexpression of miR-429 plays an important role in epithelial–mesenchymal transition28 and promotes proliferation in liver cancer cells.29 E-cadherin is a vital component of adheren junction, which is crucial in the maintenance of epithelial integrity.30 As a tumor suppressor gene, loss of E-cadherin expression is a fundamental event in tumorigenesis.31 Many studies focus on the aberrant expression of E-cadherin32,33 and discover that Snail and ZEB families are the major transcriptional repressors in downregulating E-cadherin.34,35 The Par genes (Par1, 3, 4, 5, 6, and aPKC) encode an evolutionarily conserved group of polarity proteins that is essential for cell polarization and epithelial organization.36 Par3 is a key component in delivering aPKC to the apical surface through binding of Par3 to the adapter protein Par6, which forms a constitutive complex with aPKC.37 Loss of Par3 and aPKC causes epithelial misorganization and activation of Stat3 and induction of MMP9 expression.38 It has been reported that loss of polarity proteins that are essential to epithelial polarity play a crucial role in carcinogenesis.39 E-cadherin and Par3 may act as suppressors of tumorigenesis in various cancers. Immunohistochemistry and Western blotting analysis showed that the expression levels of E-cadherin and Par3 were remarkably downregulated in colon cancer specimens, compared with those of normal colon tissues (Figure 2). However, miR-429 was significantly upregulated in cancer tissues. There might be a negative correlation between these polarity proteins and miR-429.
However, upregulated expression of miR-429 was reversed after the treatment of EVO and BER. As observed in our data, the expression level of miR-429 was significantly lower in tumor tissues than in normal tissues after treatment with BER and EVO for 72 hours, indicating that BER and EVO might exert antitumor effect by regulating the downregulating level of miR-429. BER and EVO can downregulate the expression of oncogenes to suppress the development of CRC. Previous studies reported that BER could inhibit the proliferation and migration of cancer cells by inhibiting Wnt signaling pathway.40 Furthermore, BER can suppress tumor development by inducing apoptosis.41 Many studies have shown that BER was therapeutic to CRC.42–44 EVO can inhibit cell proliferation and induce apoptosis in several kinds of cells as well.45–47 EVO can increase cleaved caspase-3 and PARP proteins and caspase-3 activity by apoptosis in COLO205 and HT-29 cells.48
Taken together, our data suggest that the levels of E-cadherin and Par3 may be downregulated due to loss of polarity during the process of tumorigenesis and metastasis of human CRC, and miR-429 might function to suppress the progression of human CRC by downregulating the expression of E-cadherin and Par3. BER and EVO may be the promising antitumor agents for human CRC by downregulating the expression of miR-429.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. | ||
Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. | ||
Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. | ||
Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–230. | ||
Liu G, Jiang C, Li D, Wang R, Wang W. MiRNA-34a inhibits EGFR-signaling-dependent MMP7 activation in gastric cancer. Tumour Biol. 2014;35:9801–9806. | ||
Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. | ||
Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs inhuman lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. | ||
Song YX, Yue ZY, Wang ZN, et al. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Cancer. 2011;10:1. | ||
Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. | ||
Ikenaga NI, Ohuchida K, Mizumoto K, et al. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann Surg Oncol. 2010;17(12):3120–3128. | ||
Mori Y, Ishiguro H, Kuwabara Y, et al. MicroRNA-21 induces cell proliferation and invasion in esophageal squamous cell carcinoma. Mol Med Report. 2009;2(2):235–239. | ||
Wu WK, Law PT, Lee CW, et al. MicroRNA in colorectal cancer: from benchtop to bedside. Carcinogenesis. 2011;32(3):247–253. | ||
Amripal S, Sanjiv D, Navpreet K, et al. Berberine: alkaloid with wide spectrum of pharmacological activities. J Nat Prod. 2010;3:64–75. | ||
Abd El-Wahab AE, Ghareeb DA, Sarhan EE, Abu-Serie MM, El Demellawy MA. In vitro biological assessment of Berberis vulgaris and its active constituent, berberine: antioxidants, anti-acetylcholinesterase, anti-diabetic and anticancer effects. BMC Complement Altern Med. 2013;13:218. | ||
Qadir SA, Kwon MC, Han JG, et al. Effect of different extraction protocols on anticancer and antioxidant activities of Berberis koreana bark extracts. J Biosci Bioeng. 2009;107:331–339. | ||
Ho YT, Yang JS, Lu CC, et al. Berberine inhibits human tongue squamous carcinoma cancer tumor growth in a murine xenograft model. Phytomedicine. 2009;16(9):887–890. | ||
Domitrovic R, Jakovac H, Blagojevic G. Hepatoprotective activity of berberine is mediated by inhibition of TNF-alpha, COX-2, and iNOS expression in CCl(4)-intoxicated mice. Toxicology. 2011;280:33–43. | ||
Saha P, Sen R, Hariharan C, Kumar D, Das P, Chatterjee M. Berberine chloride causes a caspase-independent, apoptotic-like death in Leishmania donovani promastigotes. Free Radic Res. 2009;43:1101–1110. | ||
Kan SF, Yu CH, Pu HF, et al. Anti-proliferative effects of evodiamine on human prostate cancer cell lines DU145 and PC3. J Cell Biochem. 2007;101:44–56. | ||
Kobayashi Y. The nociceptive and anti-nociceptive effects of evodiamine from fruits of Evodia rutaecarpa in mice. Planta Med. 2003;69:425–428. | ||
Lee TJ, Kim EJ, Kim S, et al. Caspase-dependent and caspase-independent apoptosis induced by evodiamine in human leukemic U937 cells. Mol Cancer Ther. 2006;5:2398–2407. | ||
Hou J, Lin L, Zhou W, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19(2):232–243. | ||
Roncucci L, Pedroni M, Vaccina F, Benatti P, Marzona L, De Pol A. Aberrant crypt foci in colorectal carcinogenesis. Cell and crypt dynamics. Cell Prolif. 2000;33:1–18. | ||
Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. | ||
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. | ||
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. | ||
Uhlmann S, Zhang JD, Schwager A, et al. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010;29(30):4297–4306. | ||
Sun Y, Shen S, Liu X, et al. Mir-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol Cell Biochem. 2014;390(1–2):19–30. | ||
Huang XY, Yao JG, Huang HD, et al. MicroRNA-429 modulates hepatocellular carcinoma prognosis and tumorigenesis. Gastroenterol Res Pract. 2013;2013:804128. | ||
Perez-Moreno M, Jamora C, Fuchs E. Sticky business. Orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. | ||
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. | ||
Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi SC, Yu Q. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 2011;71:3076–3086. | ||
Ren D, Minami Y, Nishita M. Critical role of Wnt5a-Ror2 signaling in motility and invasiveness of carcinoma cells following Snail-mediated epithelial-mesenchymal transition. Genes Cells. 2011;16:304–315. | ||
Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. | ||
Nieto MA. The snail superfamily of zinc finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. | ||
McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell. 2012;22(5):601–614. | ||
McCaffrey LM, Macara IG. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 2009;23:1450–1460. | ||
Kojima Y, Akimoto K, Nagashima Y, et al. The overexpression and altered localization of the atypical protein kinase C lambda/iota in breast cancer correlates with the pathologic type of these tumors. Hum Pathol. 2008;39:824–831. | ||
St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. | ||
Zhang J, Cao H, Zhang B, et al. Berberine potently attenuates intestinal polyps growth in ApcMin mice and familial adenomatous polyposis patients through inhibition of Wnt signalling. J Cell Mol Med. 2013;17(11):1484–1493. | ||
Kulkarni SK, Dhir A. Possible involvement of L-arginine-nitric oxide (NO)-cyclic guanosinemonophosphate (cGMP) signaling pathway in the antidepressant activity of berberine chloride. Eur J Pharmacol. 2007;569:77–83. | ||
Fukuda K, Hibiya Y, Mutoh M, Koshiji M, Akao S, Fuijwara H. Inhibition by berberine of cyclooxygenase-2 transcriptional activity in human colon cancer cells. J Ethnopharmacol. 1999;66:227–233. | ||
Fukutake M, Yokota S, Kawamura H, et al. Inhibitory effect of Coptidis Rhizoma and Scutellariae Radix onazoxymethane-induce aberrant crypt foci formation in rat colon. Biol Pharm Bull. 1998;21:814–817. | ||
Thirupurasundari CJ, Padmini R, Devaraj SN. Effect of berberine on the antioxidant status, ultrastructural modifications and protein bound carbohydrates in azoxymethane-induce colon cancer in rats. Chem Biol Interact. 2009;177:190–195. | ||
Huang YC, Guh JH, Teng CM. Induction of mitotic arrest and apoptosis by evodiamine in human leukemic T-lymphocytes. Life Sci. 2004;75:35–49. | ||
Zhang C, Fan X, Xu X, et al. Evodiamine induces caspase-dependent apoptosis and S phase arrest in human colon lovo cells. Anticancer Drugs. 2010;21:766–776. | ||
Wang KL, Hsia SM, Yeh JY, et al. Anti-proliferative effects of evodiamine on human breast cancer cells. PLoS One. 2013;8:e67297. | ||
Chien CC, Wu MS, Shen SC, et al. Activation of JNK contributes to evodiamine-induced apoptosis and g2/m arrest in human colorectal carcinoma cells: a structure-activity study of evodiamine. PLoS One. 2014;9(6):e99729. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.