Back to Journals » Cancer Management and Research » Volume 12
Effect and Mechanism of miR-26a-5p on Proliferation and Apoptosis of Hepatocellular Carcinoma Cells
Authors Zhu WJ, Yan Y, Zhang JW, Tang YD, Han B
Received 7 November 2019
Accepted for publication 22 January 2020
Published 30 April 2020 Volume 2020:12 Pages 3013—3022
DOI https://doi.org/10.2147/CMAR.S237752
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ahmet Emre Eşkazan
Wen-Jing Zhu,1 Ying Yan,2 Jiu-Wei Zhang,1 Yan-Dong Tang,3 Bo Han4
1Abdominal Ultrasonic Department, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province, People’s Republic of China; 2Oncology Department, The First Hospital of Harbin, Harbin, Heilongjiang Province, People’s Republic of China; 3Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences, Harbin, Heilongjiang Province, People’s Republic of China; 4Oncology Department, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province, People’s Republic of China
Correspondence: Bo Han
Oncology Department, The First Affiliated Hospital of Harbin Medical University, 23 Post Street, Harbin 150001, Heilongjiang Province, People’s Republic of China
Email [email protected]
Aim: This study aimed to investigate the effect and mechanism of miR-26a-5p on proliferation and apoptosis of hepatocellular carcinoma (HCC) cells.
Methods: RT-PCR was used to analyze the expression of miR-26a-5p in HCC cells and its targeted gene HMGA2 mRNA determined by biological information prediction. The rate of proliferation, invasion, apoptosis, and expression levels of related proteins of HCC cells overexpressing miR-26a-5p or those after knocking down HMGA2 expression were detected by MTT, invasion and apoptosis rate tests. Moreover, the apoptosis-promoting protein bax was upregulated and the anti-apoptosis-related protein Bcl-2 was downregulated.
Results: RT-qPCR results showed that the level of miR-26a-5p was downregulated in HCC tissues and cells, and the expression of HMGA2 was upregulated; besides, the expression of miR-26a-5p and HMGA2 was negatively correlated; miR-26a-5p was correlated with tumor diameter, differentiation degree, TNM staging and lymph node metastasis. Cell tests confirmed that miR-26a-5p functioned in tumor suppression, including inhibiting cell proliferation and invasion in two hepatocellular carcinoma cell lines and promoting apoptosis. Bioinformatics prediction and subsequent experiments proved that HMGA2 was the direct target of miR-26a-5p; moreover, after knocking down HMGA2 expression in HCC cells, cell proliferation and invasion ability were significantly inhibited, and apoptosis rate increased significantly.
Conclusion: miR-26a-5p can inhibit the proliferation and invasion of HCC cells and promote their apoptosis by directly targeting HMGA2. Abnormal decrease of miR-26a-5p and increase of its target HMGA2 are important factors that may participate in the occurrence and development of HCC. miR-26a-5p may be a new potential target for its treatment.
Keywords: miR-26a-5p, hepatocellular carcinoma cells, proliferation, apoptosis, mechanism research
Introduction
Hepatoma is one of the most common malignant tumors in the world, with high morbidity and mortality.1 However, hepatocellular carcinoma (HCC) is the most common pathological type of primary hepatoma, and about 80% of hepatoma patients are HCC.2 At present, liver transplantation and surgical resection are still the main methods for the treatment of HCC. However, since there are no obvious symptoms in the early stage of HCC, when patients show symptoms and go to the hospital, they are often in the late stage, and many of them even lose the chance of surgery.3,4 Therefore, it is of great clinical significance for HCC patients to elaborate on the biological mechanism of HCC pathogenesis and find new therapeutic targets.
MicroRNA (miRNA), as an endogenous non-coding single-stranded RNA, usually regulates the expression of target genes by combining with the 3ʹ-untranslated region (3ʹ UTR end) of the target genes, which plays a very important role in the occurrence and development of tumors.5 In recent years, with the development of bioinformatics, the role of miRNA in HCC has been paid more and more attention. miR-26a-5p is one of the most popular miRNA in recent years. miRNA has low expression in various tumors such as colorectal cancer and prostate cancer, and it is believed that miRNA may play a tumor-suppressor gene role.6 It has been reported7 that miR-26a-5p can inhibit the proliferation, invasion and metastasis of papilloma thyroid cells by inhibiting Wnt5a. In addition, some studies8 have found that miR-26a-5p can promote the development of bladder cancer by targeting PTEN. However, the role of miR-26a-5p in HCC and its related mechanisms have not been explored yet. We found that miR-26a-5p and high mobility group protein A2 (HMGA2) have targeted relationship through our detection of dual-luciferase report, and HMGA2 is a protein known to promote cancer cell proliferation and metastasis in a variety of tumors, including liver cancer,9 For example, some studies have found that it can promote gastric cancer cell metastasis and epithelial–mesenchymal transition.10 Moreover, HMGA protein, as a structural factor required for chromosome structure, plays a very important part in the occurrence and development of tumors.11
In order to further explore the proliferation, apoptosis and its mechanism of miR-26a-5p on hepatocellular carcinoma cells, we have carried out the following research in order to provide new molecular targets and more sufficient theoretical basis for HCC treatment.
Materials and Methods
Clinical Data
Seventy-six patients with liver cancer who underwent hepatic carcinectomy in our hospital from July 2013 to July 2016 were selected as a research group, including 45 male and 31 female patients. They were (56.81±3.44) years old on average. Seventy-six cases of hepatocellular carcinoma tissue and 76 cases of paracancerous tissue were obtained during the operation with the consent of the patients. Inclusion criteria: patients with liver cancer confirmed by pathological diagnosis and those with estimated survival greater than 3 months were included in the research group. Exclusion criteria were as follows: patients with other malignant tumors; patients who received any treatment before the experiment; patients with infection, blood or immune system disorders. All patients and their families agreed to participate in the experiment and signed an informed consent. This experiment was approved by the hospital ethics committee.
Experimental Materials
Human hepatocellular carcinoma cell lines HepG2, SMMC-7721, MHCC-97H, Huh-7 and human normal hepatocyte line HL-7702 were all purchased from Shanghai Institute of Cell Biology, and fetal bovine serum (FBS) and PBS were all purchased from Gibco Company, USA (10437028, 10010010,049). Trypsin, RIPA, BCA protein kits were all purchased from Thermo Scientific Company (A40007, 89901, 23229). Dual-luciferase report gene detection kit was from Solarbio Company, Beijing, China, D0010; Trizol reagent was from Applied Invitrogen, USA; qPCR kit and minScript reverse transcription kit were from TaKaRa company, Dalian, China; Annexin V-FITC/PI apoptosis kit was from Jiangsu kaiji Biology Co., Ltd. HMGA2, Bax, Bcl-2 and β-actin primary antibody and goat anti-rabbit secondary antibody were all purchased from Shanghai Unionville Company.
Rt-PCR Detection of miR-26a-5p and HMGA2 Expression
The expression levels of miR-26a-5p and HMGA2 in HepG2, SMMC-7721, MHCC-97H, Huh-7, and human normal hepatocyte line HL-7702 were detected. First, the cell line was prepared into cell suspension. Then, TRIzol reagent was added to extract the total RNA. The purity and concentration of the extracted total RNA were tested with an ultraviolet spectrophotometer. SYBR-Green Realtime PCR Master mix was used to reversely transcribe the total RNA with miR-26a-5p and HMGA2. The operation steps were strictly in accordance with the manufacturers’ kit. Then, PCR amplification experiment was carried out. PCR reaction system was as follows: cDNA 1 μL, upstream and downstream primers 0.4 μL, 2×SYBRr-Green Realtime PCR Master mix 10 μL, Passive Reference Dye (50×) 0.4 μL, ddH2O added to 20 μL. The PCR reaction conditions of miR-26a-5p were as follows: Pre-denaturation at 94°C for 45 s, denaturation at 94°C for 10 s, annealing at 60°C for 30 s, with a total of 40 cycles; the experiment was carried out three times, using U6 as an internal reference. HMGA2 reaction conditions were as follows: Denaturation at 94°C for 10 s, annealing at 60°C for 30 s, with a total of 40 cycles; the experiment was carried out three times, using β-actin as internal reference. miR-26a-5p upstream sequence was as follows: 5ʹ-ATGGTTCGTGGGTTCAAGTAATCGATGGC-’3; downstream sequence was as follows: 5ʹ-GCAGGGTCCGAGTATTCG-’3; U6 upstream sequence was as follows: 5ʹ-CTCGCTTCGGCACA-’3, downstream sequence 5ʹ-AACCTTCACGAATTCGT-’3; HMGA2 was as follows: upstream sequence: 5ʹ-AAAGCAGAAAGCCACGAAA-3ʹ, downstream sequence: 5ʹ-TTCCTCCGAGAGCTTCTT-3ʹ; β-actin, upstream sequence: 5ʹ-GATCATTGCTCCTCCTGAGC-3ʹ, downstream sequence: 5ʹ-ACTCCTGCTTGCTGATCCAC-3ʹ.
Transfection of Cell Line
By detecting the expression levels of miR-26a-5p and HMGA2 in cell lines, it was found that the expression of miR-26a-5p in HepG2 and SMMC-7721 cell lines was lower than that in cell lines of the other two groups. Therefore, these two cell lines were selected for transfection and subsequent experiments. miR-26a-5p mimetic, miRNA mimetic negative control (miR-NC), small interfering RNA (siRNA) (si-HMGA2) of HMGA2 and its negative control (si-NC) were produced by GenePharma (Shanghai, China). Cell transfection was carried out according to the manufacturers’ protocol using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
MTT Measurement
MTT measurement (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used to evaluate cell viability. HepG2 and SMMC-7721 cells transfected with miRNA mimetic or siRNA only or co-transfected with miRNA mimetic and HMGA2 plasmid were inoculated into 96-well plates at a density of 3×103 cells per well, and 20μL of MTT (0.5mg/mL) solution was added to each well when incubated at 37°C for 24 h, 48 h and 72 h, and incubation was continued at 37°C for 3 h. Subsequently, 250uL of dimethyl sulfoxide was added to each well, and finally, absorbance at 490 nm was measured by ELISA (Molecular Devices, LLC, Sunnyvale, CA, USA).
Measurement of Cell Invasion
First, Matrigel matrix glue (BD Biosciences, San Jose, CA, USA) was diluted at a ratio of 1:8 and then coated with the upper chamber surface of the bottom membrane of the Transwell chamber (8μm pore size; BD Biosciences, San Jose, CA, USA). The upper chamber surface of the bottom membrane was placed at 37°C for 30 min to polymerize Matrigel into gel, and the bottom membrane was hydrated before use. The transfected cells were starved for 24 h, then the cells were resuspended with DMEM culture solution without FBS and the cell density was adjusted to 3×104 cells/mL. The cells were then inoculated in the upper chamber of Transwell of the 24-well plate, about 200 µL of cell suspension was added to each chamber, 600 μL of DMEM culture solution containing 10%FBS was added to the lower chamber of the 24-well plate, and then cultured in an incubator at 37°C for 48 h. After the culture was completed, the supernatant was removed with cotton swabs, and then the removed chambers were washed with PBS. Cells in the lower chamber were fixed with 95% ethanol solution for 15 min. After taking out, they were washed again with PBS, and then stained with 0.5% crystal violet. After staining, the migration number of cells in 6 random wells was calculated randomly with a microscope, and the average value was calculated. The experiment was repeated 3 times.
Apoptosis Detection
Annexin V-FITC/PI double staining combined with flow cytometry was used to detect apoptosis. Transfected cells were inoculated at a density of 3×105 cells/well in a 6-well plate, digested with 0.25% trypsin, washed twice with PBS after digestion, added with 100μL of binding buffer, prepared into 1x106 cells/mL suspension, sequentially added with AnnexinV-FITC and PI, incubated in dark at room temperature for 5 min, and detected with FC500MCL flow cytometer system. The experiment was repeated for 3 times and averaged.
Gene Measurement of Dual Luciferase Report
It was performed to determine whether HMGA2 was a direct target gene of miR-26a-5p. HMGA2 3ʹUTR dual-luciferase report plasmids (WT and MUT) were constructed by RiboBio. They were co-transfected into HepG2 or SMMC-7721 cells with miR-26a-5p mimetic or mimetic control by Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific, Inc., USA). Forty-eight hours after incubation, luciferase activity was detected by a dual-luciferase determination system (Promega Corp, Madison, WI, USA).
Western Blot Detection
After collecting cells, the RIPA lysis method was used to extract total protein, and BCA method was used to detect the protein concentration; the protein concentration was adjusted to 4μg/μL, and 12% SDS-PAGE electrophoresis separation was carried out. After ionization, the membrane was transferred to the PVDF membrane, stained with Lichun red working solution, soaked in PBST for 5 min, washed and sealed with 5% defatted milk powder for 2 h. And after membrane transfer, a monoclonal antibody against HMGA2 (1:500), Bax (1:500), Bcl-2 (1:500) and β-actin (1:1000) was added to seal overnight at 4°C. The membrane was washed to remove the primary antibody, and horseradish peroxidase-labeled goat anti-rabbit secondary antibody (1: 5000) was added, incubated at 37°C for 1 h and rinsed with PBS for 3 times; the excess liquid was absorbed after rinsing, and then ECL emitted light and developed.
Statistical Methods
In this study, SPSS 19.0 software was used to analyze the data, GraphPad Prism 6 software was used to draw relevant pictures, the measurement data were expressed by mean±standard deviation (SD±meas), t-test was used, independent sample t-test was used for inter-group comparison that was expressed by t, one-way analysis of variance was used for multi-group comparison, LSD-t-test was used for post-event two-two comparison, and Pearson was used for correlation analysis. A p value lower than 0.05 was considered to be statistically different.
Results
Expression Levels of miR-26a-5p and HMGA2 in HCC and Clinical Significance
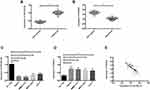
QRT-PCR was used to detect the expression levels of miR-26a-5p and HMGA2 in HCC tissue, four HCC cell lines and one human normal hepatocyte line HL-7702. The results showed that the expression of miR-26a-5p in HCC was significantly lower than that in paracancerous tissue (P < 0.05), and the expression of HMGA2 mRNA was significantly higher than that in paracancerous tissue (P < 0.05); the expression of miR-26a-5p in HCC cell line was lower than HL-7702 (P < 0.05), and the expression of HMGA2 in HCC cell line was higher than HL-7702 (P < 0.05). Through correlation analysis found that the expression of miR-26a-5p and HMGA2 was negatively correlated (P < 0.05) in HCC tissue. According to the expression of miR-26a-5p on average, the patients were divided into high and low expression groups. Further analysis of the relationship between miR-26a-5p and pathological data showed that miR-26a-5p was closely related to tumor diameter, differentiation degree, TNM staging and lymph node metastasis (P < 0.05). (Figure 1, Table 1).
 |
Table 1 Relationship Between miR-26a-5p and Pathological Data of Patients with Liver Cancer |
Effects of miR-26a-5p Overexpression on Proliferation, Invasion and Apoptosis of HCC Cells
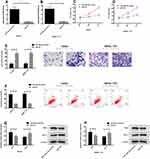
In order to study the effects of miR-26a-5p overexpression on the proliferation, invasion and apoptosis of HCC cells, HepG2 and SMMC-7721 cells were transfected with miR-26a-5p mimetic or miR-NC. Forty-eight hours after transfection, compared with miR-NC, miR-26a-5p in HepG2 and SMMC-7721 cells transfected with miR-26a-5p mimetic increased significantly (Figure 2A and B, P < 0.05). The results showed that miR-26a-5p mimetics were effective as endogenous miR-26a-5p in vitro. In addition, MTT measurement, cell invasion and flow apoptosis measurement were performed after transfection. As shown in Figure 2C–H, the overexpression of miR-26a-5p inhibited proliferation (P < 0.05) and invasion (P < 0.05) of HepG2 and SMMC-7721 cells significantly, and so was their apoptosis. What’s more, WB experiments showed that the expression of pro-apoptotic protein bax increased and that of anti-apoptotic protein Bcl-2 decreased in cells overexpressed by miR-26a-5p, as shown in Figure 2G–H.
HMGA2 Is the Direct Target Gene of miR-26a-5p
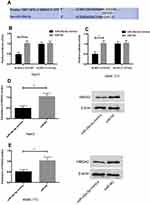
In order to explore the potential mechanism of miR-26a-5p in HCC, bioinformatics analysis was conducted to predict the target gene of miR-26a-5p. HMGA2 was identified as a target gene for miR-26a-5p (Figure 3A). In order to check whether the 3ʹ UTR of HMGA2 can be directly targeted by miR-26a-5p, luciferase report gene measurement was carried out. The results showed that (Figure 3B and c) the overexpression of miR-26a-5p decreased luciferase activity of position 1867–1873 of HMGA2 3ʹ UTR Mut (P < 0.05), but had no effect on position 1867–1873 of HMGA2 3ʹ UTR Mut; Western blot showed that the expression of HMGA2 protein in miR-26a-5p simulated transfected HepG2 and SMMC-7721 cells was downregulated (Figure 3D and E, P <0.05).
Effects of Downregulation of HMGA2 Expression on Proliferation, Invasion and Apoptosis of HCC Cells
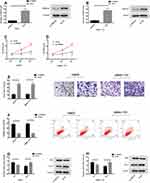
In order to confirm whether the tumor inhibitory effect of miR-26a-5p in HCC was mediated by HMGA2, we knocked down the expression of HMGA2 in HepG2 and SMMC-7721 cells. Western blot analysis showed that after transfection with si-HMGA2, HMGA2 was downregulated in HepG2 and SMMC-7721 cells (Figure 4A and B, P < 0.05). After transfection, MTT measurement and cell invasion measurement si-HMGA2 inhibited the proliferation and invasion of HepG2 and SMMC-7721 cells and promoted their apoptosis (Figure 4C–F, P < 0.05) significantly. Besides, WB experiment showed that the expression of pro-apoptotic protein bax increased and the expression of anti-apoptotic protein Bcl-2 decreased in cells knocked down HMGA2 expression (Figure 4G–H). These results further indicated that HMGA2 was the direct functional target of miR-26a-5p in HCC.
Discussion
In recent years, the mechanism of miRNA in cancer has gradually become a hot topic of research, and targeted therapy is also a popular treatment method that arises at the historic moment.12 In addition, a large number of studies13,14 have shown that miRNA is closely related to the occurrence and development of HCC, and believed that the imbalance of various miRNA and genes is one of its main causes.
In this study, we found that the expression of miR-26a-5p was downregulated both in HCC tissue and cell line. Previous studies15 have also reported that miR-26a-5p, as a tumor suppressor, can block the cycle of prostate cancer and apoptosis by regulating its target; other studies16 have pointed out that miR-26a-5p can promote the metastasis of human lung cancer cells by activating JAK2/STAT3 axis, which may play a role of oncogene in lung cancer. HCC is the most common malignant tumor with high mortality17 currently, and our discovery provides important evidence for discovering potential therapeutic targets of HCC cells. Previous reports on miR-26a-5p have indicated that miR-26a-5p mainly plays the role of tumor-suppressor gene.18 However, we also observed the effect of miR-26a-5p on HCC cells by regulating its overexpression. The results showed that when we upregulated the expression of miR-26a-5p, the proliferation and invasion ability of HCC cells were obviously inhibited, and the apoptosis rate also increased obviously, which indicated that miR-26a-5p might also play a role of tumor-suppressor gene in HCC. A previous study19 upregulated miR-26a expression in a hepatoma mouse model by adenovirus and found that the proliferation of HCC cells was inhibited. There was also research20 indicated that, when exploring the action mechanism of miR-26a-5p in lung adenocarcinoma, miR-26a-5p could inhibit the occurrence of tumors in vivo by regulating the expression level of E2F7. The above researches can confirm our conclusion.
The above results show the effect of miR-26a-5p on HCC cells, but the specific mechanism of action has not been discussed in detail yet. Through bioinformatics prediction, we found that HMGA2 was the direct target of miR-26a-5p, and the expression of HMGA2 protein in HCC cells transfected by miR-26a-5p simulation had been downregulated. HMGA2 is a kind of transcription factor, belonging to the family of high mobility genes. It is a protein known to promote the proliferation and metastasis of cancer cells in various tumors including hepatoma.21 Our results showed that HMGA2 was highly expressed in HCC tissues and cells, and the expression levels of HMGA2 and miR-26a-5p were negatively correlated in HCC tissues, which also provided important evidence for the correlation between them. Subsequently, in order to further confirm that the role of miR-26a-5p in hepatoma was mediated by HMGA2, we knocked down the expression of HMGA2 in HCC cells, and then observed the biological function of them; the results showed that when the expression of HMGA2 was knocked down, the proliferation and invasion ability of HCC cells were significantly inhibited, and the apoptosis rate was increased significantly. The above conclusions can fully prove that HMGA2 is an important target of miR-26a-5p in HCC cells. Regarding the mechanism of action of HMGA2, previous studies22 have shown that HMGA2 is an important target for chromosomal abnormalities. One of its mechanisms for promoting tumor progression is that the expression of HMGA2 after chromosome truncation has deleted the carboxyl terminal of the 3ʹ non-coding region, which leads to the fusion of HMGA2 with some tumor-suppressor genes and loss of tumor-suppressor functions of these tumor-suppressor genes, thus promoting the development of tumors. Another study9 has indicated that HMGA2 may promote tumor invasion and metastasis through epithelial–mesenchymal transition. For example, previous studies23 have found that HMGA2 can induce epithelial–mesenchymal transition in pancreatic cancer by conditioning the Ras pathway. In addition, it has been clearly pointed out in another study24 that silencing HMGA2 expression may affect the progression of colorectal cancer by inducing apoptosis and cell cycle arrest of cells, which also proves that HMGA2 has a certain role in apoptosis of cancer cells, which can confirm our conclusion.
To sum up, miR-26a-5p inhibits the proliferation and invasion of HCC cells and promotes their apoptosis by directly targeting HMGA2. Abnormal decrease of miR-26a-5p and increase of its target HMGA2 are important factors that may participate in the occurrence and development of HCC. This novel miR-26a-5p/HMGA2 axis provides an in-depth understanding of the pathogenesis of HCC and may be a new potential target for its therapy. However, there are still some deficiencies in this study. For example, on the one hand, the specific mechanism of action of miR-26a-5p in HCC is simply described by quoting relevant documents, and no detailed experimental investigation has been carried out; on the other hand, whether the role of miR-26a-5p in HCC regulates other genes besides HMGA2 remains to be further tested.
Acknowledgment
This study (2016LCZX70) was supported by the Innovative Scientific Research Project of Harbin Medical University, American Ginseng Polysaccharide PPQ inhibits the growth of Hepatocellular carcinoma cells by inhibiting the expression of AEG-1.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
2. Omata M, Cheng AL, Kokudo N, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017;11(4):317–370.
3. Yang X, Zhang D, Liu S, Li X, Hu W, Han C. KLF4 suppresses the migration of hepatocellular carcinoma by transcriptionally upregulating monoglyceride lipase. Am J Cancer Res. 2018;8(6):1019–1029.
4. Jiang T, Guan LY, Ye YS, Liu HY, Li R. MiR-874 inhibits metastasis and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting SOX12. Am J Cancer Res. 2017;7(6):1310–1321.
5. Tao J, Liu Z, Wang Y, et al. MiR-542-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting UBE3C. Biomed Pharmacother. 2017;93:420–428. doi:10.1016/j.biopha.2017.06.070
6. Ghanbari R, Mosakhani N, Asadi J, et al. Downregulation of plasma MiR-142-3p and miR-26a-5p in patients with colorectal carcinoma. Iran J Cancer Prev. 2015;8(3):e2329. doi:10.17795/ijcp
7. Shi D, Wang H, Ding M, et al. MicroRNA-26a-5p inhibits proliferation, invasion and metastasis by repressing the expression of Wnt5a in papillary thyroid carcinoma. Onco Targets Ther. 2019;12:6605–6616. doi:10.2147/OTT.S205994
8. Wang H, Hu Z, Chen L. Enhanced plasma miR-26a-5p promotes the progression of bladder cancer via targeting PTEN. Oncol Lett. 2018;16(4):4223–4228. doi:10.3892/ol.2018.9163
9. Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283(48):33437–33446. doi:10.1074/jbc.M802016200
10. Dong J, Wang R, Ren G, et al. HMGA2-FOXL2 axis regulates metastases and epithelial-to-mesenchymal transition of chemoresistant gastric cancer. Clin Cancer Res. 2017;23(13):3461–3473. doi:10.1158/1078-0432.CCR-16-2180
11. Zhao YC, Jiao Y, Li YQ, Fu Z, Yang ZY, He M. Elevated high mobility group A2 expression in liver cancer predicts poor patient survival. Rev Esp Enferm Dig. 2019;112.
12. Shi C, Zhang Z. MicroRNA-362 is downregulated in cervical cancer and inhibits cell proliferation, migration and invasion by directly targeting SIX1. Oncol Rep. 2017;37(1):501–509. doi:10.3892/or.2016.5242
13. Xu Q, Liu X, Liu Z, et al. MicroRNA-1296 inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting SRPK1-mediated PI3K/AKT pathway. Mol Cancer. 2017;16(1):103. doi:10.1186/s12943-017-0675-y
14. Yuan J, Ji H, Xiao F, et al. MicroRNA-340 inhibits the proliferation and invasion of hepatocellular carcinoma cells by targeting JAK1. Biochem Biophys Res Commun. 2017;483(1):578–584. doi:10.1016/j.bbrc.2016.12.102
15. Rizzo M, Berti G, Russo F, et al. Discovering the miR-26a-5p targetome in prostate cancer cells. J Cancer. 2017;8(14):2729–2739. doi:10.7150/jca.18396
16. Song Q, Liu B, Li X, et al. MiR-26a-5p potentiates metastasis of human lung cancer cells by regulating ITGbeta8- JAK2/STAT3 axis. Biochem Biophys Res Commun. 2018;501(2):494–500. doi:10.1016/j.bbrc.2018.05.020
17. Bao H, Li X, Li H, et al. MicroRNA-144 inhibits hepatocellular carcinoma cell proliferation, invasion and migration by targeting ZFX. J Biosci. 2017;42(1):103–111. doi:10.1007/s12038-016-9662-5
18. Chang L, Li K, Guo T. miR-26a-5p suppresses tumor metastasis by regulating EMT and is associated with prognosis in HCC. Clin Transl Oncol. 2017;19(6):695–703. doi:10.1007/s12094-016-1582-1
19. Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi:10.1016/j.cell.2009.04.021
20. Liang R, Xiao G, Wang M, et al. SNHG6 functions as a competing endogenous RNA to regulate E2F7 expression by sponging miR-26a-5p in lung adenocarcinoma. Biomed Pharmacother. 2018;107:1434–1446. doi:10.1016/j.biopha.2018.08.099
21. Wang Y, Chen F, Zhao M, et al. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J Biol Chem. 2017;292(37):15395–15407. doi:10.1074/jbc.M117.783738
22. Sgarra R, Pegoraro S, Ros G, et al. High Mobility Group A (HMGA) proteins: molecular instigators of breast cancer onset and progression. Biochim Biophys Acta Rev Cancer. 2018;1869(2):216–229. doi:10.1016/j.bbcan.2018.03.001
23. Gong J, Wang Y, Jiang B, Xu B, Hu C. Impact of high-mobility-group A2 overexpression on epithelial-mesenchymal transition in pancreatic cancer. Cancer Manag Res. 2019;11:4075–4084. doi:10.2147/CMAR.S199289
24. Esmailzadeh S, Mansoori B, Mohammadi A, Shanehbandi D, Baradaran B. siRNA-mediated silencing of HMGA2 induces apoptosis and cell cycle arrest in human colorectal carcinoma. J Gastrointest Cancer. 2017;48(2):156–163. doi:10.1007/s12029-016-9871-z
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.