Back to Journals » OncoTargets and Therapy » Volume 13
Dysregulation of SPRR3/miR-876-3p Axis Contributes to Tumorigenesis in Non-Small-Cell Lung Cancer
Authors Li Q, Wang Y, Hu R , Yang G
Received 9 January 2020
Accepted for publication 14 March 2020
Published 23 March 2020 Volume 2020:13 Pages 2411—2419
DOI https://doi.org/10.2147/OTT.S245422
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Qin Li,1,* Yuxuan Wang,2,* Rongkuan Hu,3 Guang Yang1
1Department of Oncology, BenQ Medical Center, Nanjing Medical University, Nanjing 210029, People’s Republic of China; 2Department of Thoracic Surgery, The First Affiliated Hospital of Soochow University, Suzhou, 215006, People’s Republic of China; 3GenePharma Co., Ltd, Suzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Rongkuan Hu 199 Dongping Street, Suzhou 215123, People’s Republic of China
Tel +86-512-86668828
Email [email protected]
Guang Yang 181 Zhuyuan Road, Suzhou 215010, People’s Republic of China
Tel +86-512-80838800
Email [email protected]
Background: SPRR3, also known as esophagin, has been shown to be involved in the initiation and progression of numerous types of tumor. However, the biological function of SPRR3 that contributes to non-small-cell lung cancer (NSCLC) growth and migration is largely unknown.
Methods: The expression of SPRR3 and its association with EZH2 and miR-876-3p in NSCLC cells were determined by real-time PCR. Protein levels were measured by immunohistochemistry (IHC) and Western blot. Cell functions were studied by CCK-8, transwell assay, flow cytometry and dual-luciferase reporter assay. The effect of SPRR3 on tumor growth in vivo was evaluated in patient-derived xenograft (PDX) models.
Results: SPRR3 was up-regulated in most NSCLC cell lines and clinical tissues. Also, the correlation between SPRR3 expression and clinical features was significant. Functional studies confirmed that SPRR3 modulates cell proliferation, invasion and cell apoptosis in NSCLC via regulating EZH2, which is a well-known oncogene in NSCLC. Furthermore, SPRR3 was found to be a direct target of miR-876-3p that also plays a suppressor role in NSCLC.
Conclusion: These findings indicated that miR-876-3p/SPRR3/EZH2 signaling cascade exerts important roles in the regulation of NSCLC, suggesting that this pathway can serve as a potential therapeutic target in NSCLC.
Keywords: SPRR3, EZH2, miR-876-3p, NSCLC, tumorigenesis
Introduction
Lung & bronchus cancers are a set of lethal diseases, with estimated new cases about 234,030 and killed almost 154,050 Americans in 2018.1 Lung cancer mainly includes two subtypes based on pathological differences, including non-small-cell lung cancer (NSCLC) and small cell lung cancer (SCLC).2 Metastasis is the primary cause of death in NSCLC. Herein, it is important to predict the possibility of occurrence at early stages and thereby cured by surgery, targeted medicine or immune therapy. Besides, several studies indicated that the complicated micro-environments of NSCLC may contribute to the metastasis and can be served as diagnostic and therapeutic targets.2,3
In a previously study, we performed mass spectrometry based secretome methods and characterized the micro-environmental factors that may be involved in the metastatic roles of NSCLC. As described information, we found six genes, including SPRR3, play roles in NSCLC cell metastasis and survival.4
Among those proteins, SPRR3, a member of the large family of the small proline-rich proteins (SPRRs), is located within the epidermal differentiation complex on chromosome 1q21.5 High expression of SPRR3 is associated with cancer carcinogenesis in glioblastoma multiform, colorectal and breast cancer.6–8 SPRR3 has also been reported to interact with c-Jun and Fra1 in human bronchial epithelial cells.9 Besides, SPRR3 is found to play roles in vascular smooth muscle cells (VSMCs) adaption to local biomechanical stress.10 In particular, SPRR3 was reported to promote phosphorylation of AKT in breast cancer and colorectal tumorigenesis.7,8 However, the exact role of SPRR3 in the invasive capacity of NSCLC remains unknown.
NSCLC tumorigenesis has been identified as a multiple processes including post-transcriptional regulation and genetic modifications, including a set of microRNAs (miRNAs). miRNAs are 22-nt RNAs that bind to 3ʹUTR of messenger RNA (mRNA) and regulate essential biological processes in many types of tumor. For example, miR-140-3p regulates squamous cell lung cancer (SqCLC) by targeting BRD9, while miR-342-3p modulates NSCLC tumorigenesis via suppressing AGR2.2,11 Furthermore, miRNA expression profiles are correlated with NSCLC patients’ survival and can be served as diagnostic and prognostic markers.12
In this study, we showed SPRR3 is up-regulated in NSCLC cells and tissues, which consistent with the data from the Cancer Genome Atlas (TCGA). Moreover, SPRR3 expression is correlated with tumor stage and metastasis status. Knockdown of SPRR3 suppresses cell growth, invasion and promotes apoptosis. Further investigation demonstrated that SPRR3 regulates EZH2 and is targeted by miR-876-3p. Collectively, miR-876-3p/SPRR3/EZH2 axes play roles in NSCLC tumorigenesis and can be severed as potential treatment targets.
Materials and Methods
Clinical Tumor Specimens
The cohorts of patient tissues were obtained from 2013 to 2018 in the first affiliated hospital of Soochow university and the BenQ Medical Center (Table 1). Written informed consent was obtained from all enrolled subjects, and the study was approved by the Ethics Committee of The authors’ institution, in accordance with the Declaration of Helsinki.
 |
Table 1 Clinical Features |
Materials
EZH2 (ab186006) and Actin (ab179467) primary antibodies and secondary antibodies (goat anti rabbit IgG, ab205718) were got from abcam (Cambridge, UK). SPRR3 antibody (11742-1-AP) was purchased from ProteinTech (Wuhan, China). p-Akt (ser 473)(#9271) antibody was got from Cell Signaling Technology (Danvers, MA, USA). SPRR3 siRNAs, miR-876-3p mimics and related mimics control were got from GenePharma (Table 2, Suzhou, China). Fetal Bovine Serum (FBS) and RPMI-1640 were got from Sigma-Aldrich. CCK-8 kit was purchase from Dojindo (Tokyo, Japan).
 |
Table 2 siRNA and miRNA Sequences |
Cell Culture and Transfection
All lung cell lines used here were purchased from Chinese Academy of Sciences (Shanghai, China). All NSCLC cells and Beas-2b were cultured in RPMI-1640 supplied with 10% FBS at 37°C in 5% CO2. Lipofectamine 2000 reagent was used for transfection of miRNAs following the manufacturer’s protocol (Invitrogen, Waltham, MA, USA).
Western Blotting Analysis and Immunohistochemistry Assays
Cells were harvested and subjected to 10% SDS-PAGE and transferred to nitrocellulose blotting membranes (GE Healthcare, Dallas, TX, USA). The membranes were incubated with antibodies targeting SPRR3 or EZH2 overnight at 4 °C (1:1000), following by blotted with the secondary antibody for 1 h at room temperature. Membranes were developed using standard methods previously described.4 Immunohistochemistry was followed according to the methods published previously.11
Quantitative Real-Time PCR (qRT-PCR)
RNA was extracted from clinical tissues or cells using Trizol (Invitrogen, Waltham, MA, USA). Then, SuperScript II reverse transcriptase kit (Invitrogen) and Power SYBR Green Mix (Applied Biosystems) were used for qPCR. The relative expression of RNA was measured using the inverse log of the delta-delta CT and normalized to the reference gene. The primers were used as follows: SPRR3 sense 5ʹ- GTTCATCGTGTTCGTGGCT −3ʹ, SPRR3 antisense 5ʹ- AGGGCATCATTTGAGTCCTG −3ʹ, GAPDH sense 5ʹ- CATGAGAAGTATGACAACAGCCT −3ʹ, GAPDH antisense 5ʹ- AGTCCTTCCACGATACCAAAGT −3ʹ, U6 sense, 5ʹ-ATTGGAACGATACAGAGAAGATT-3ʹ; and U6 antisense, 5ʹ-GGAACGCTTCACGAATTTG-3ʹ.
Cell Growth, Apoptosis and Invasion
Cell growth was detected by Cell counting kit following the instruction. Briefly, after transfection of siRNA or miRNAs, the absorbance of each sample was measured at 450nm. For apoptosis assay, cells were washed and stained with Annexin V-FITC Apoptosis detection kit (ThermoFisher, Waltham, MA, USA) and measured on the flow cytometer (BD Biosciences, San Jose, CA, USA). For the invasion assay, the trans-membranes were pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA). After transfection of RNA oligos, cells were starved and seeded into the upper chamber of the inserts in serum media. The low chamber was incubated with 10% serum. After 48h, the cells were fixed and stained using 0.5% crystal violet.4
Luciferase Reporter System
PC9 cells were co-transfected with wild type pGL3-SPRR3, and mutant pGL3-SPRR3 (Promega, Madison, Wisconsin, USA). MiRNA mimics or negative control (scramble) were co-transfected using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA). The cells were harvested 24 h after transfection. Dual-luciferase was detected using the Dual-Luciferase Reporter assay kit (Promega, Madison, Wisconsin, USA) by analyzing the Firefly and Renilla luciferase activities.
Animals and Ethics Statement
Fresh NSCLC tumor tissues samples (patient ID: 1290024, Male, Age 55, F0) were kept on ice and implanted subcutaneously into NOD/SCID mice (Vital River Laboratory Animal Technology, Beijing, China) as previously described. In this study, F5 PDX models with tumor volumes of 100–200 mm3 were used. Tumor growth was monitored and photographed. This study was approved by the Ethics Committee for Animal Experimentation of the Soochow University and conducted according to the National Institutes of Health guide for the care and use of laboratory animals.
Statistical Analysis
1129 lung samples according to RNA-seq data (https://tcga.xenahubs.net/download/TCGA.LUNG.sampleMap/HiSeqV2.gz) was download, and the log2 transformed expression in patient and normal control was analyzed. All data were analyzed by GraphPad Prism 6.0 and presented with mean ± SD. Significance between each groups were analyzed by student’s t-test (two groups) or one-way ANOVA (more than two groups).
Results
SPRR3 Is Up-Regulated and Correlated with Clinical Features in NSCLC
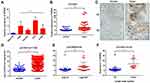
To investigate the role of SPRR3 in human NSCLC carcinogenesis, we first detected the expression of SPRR3 in 4 NSCLC cell lines, including H1299, PC9, HCC827 and A549. Beas-2b is a normal lung epithelial cell and was set as a negative control. As a result, the mRNA expression of SPRR3 was higher in almost all of these NSCLC cells compared to normal control (Figure 1A). Subsequently, we assessed the SPRR3 expression in 74 patients’ specimens using qPCR and IHC, including tumor tissues and paired adjacent normal control. As shown in Figure 1B and C, the RNA and protein level of SPRR3 were both overexpressed in tumor tissues compared to normal control. Moreover, we confirmed this finding by analyzing a cohort of 1129 lung samples according to RNA-seq data (https://tcga.xenahubs.net/download/TCGA.LUNG.sampleMap/HiSeqV2.gz). In consistent with our previous data, SPRR3 was up-regulated in tumors rather than in normal adjacent (Figure 1D).
We subsequently evaluated the relationship between SPRR3 and clinical features in our sample cohort. According to the mRNA expression level, we found SPRR3 was more likely expressed in late stage NSCLC patients (Figure 1E). Furthermore, SPRR3 expression was elevated in lymph node metastasis specimen (N1/N2) rather than in N0 patients (Figure 1F). This clue was a clinical confirmation to our previous study that SPRR3 was involved in NSCLC metastasis.4
SPRR3 Effected NSCLC Cell Growth Both in vitro and in vivo
To investigate the role of SPRR3 in NSCLC cells, cell growth was detected by CCK8 assay. Consistent with previously studies,7,8 after transfected with SPRR3 siRNAs, SPRR3 protein associated with phosphorylation of Akt levels were decreased (Figure 2A). Considering Akt modulates EZH2 expression,13 a well-known oncogene in lung cancer, we detected whether knockdown of SPRR3 affects EZH2 expression. As shown in Figure 2A, EZH2 protein level was also reduced by inhibiting SPRR3. Furthermore, SPRR3 targeted siRNAs significantly inhibited cell viability after 48h and 72h transfection in two NSCLC cell lines (Figure 2B and C). In addition, to determine whether SPRR3 is involved in tumor growth in patient-derived xenograft models, we injected SPRR3 siRNAs twice a week (2OD). As shown in Figure 2D and E, siRNAs targeting SPRR3 suppressed the tumor growth of NSCLC PDX models. These results demonstrate that inhibiting of SPRR3 suppress NSCLC cancer growth both in vitro and in vivo.
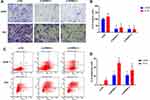
SPRR3 Regulates Cell Invasion and Apoptosis in NSCLC
As described previously, SPRR3 was involved in NSCLC cell migration, but the effect in cell invasion was not unknown.4 Toward this end, we first measured cell invasion via trans-well assay. As a result, knockdown of SPRR3 in NSCLC cell lines led an inhibition of cell invasion by more than 50% compared to the negative control in two NSCLC cell lines (Figure 3A and B). Moreover, we detected the rate of cell apoptosis after transfection of siRNAs-SPRR3 by using FACS. To our predicted, knock-down of SPRR3 promoted NSCLC cell apoptosis (Figure 3C and D). These results indicate SPRR3 is an oncogene and can be served as a treatment target.
SPRR3 Regulates EZH2 in NSCLC
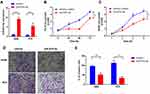
As mentioned in Figure 2A, EZH2 level was decreased after knockdown of SPRR3. EZH2 is a member of the polycomb repressive complex responsible for histone methylation and commonly overexpressed as an oncogene across several cancers, including NSCLC.13,14 To further verify the relationship between SPRR3 and EZH2 in NSCLC, we first analyzed a cohort as mentioned above of 1129 patients harboring SPRR3 and EZH2. As shown in Figure 4A, EZH2 is positively correlated with SPRR3, suggesting a linkage between them. We further restored the EZH2 expression by transfection of pLEX-EZH2 vectors in si-SPRR3 group in PC9 (Figure 4B). To our predicted, EZH2 can rescue the cell growth which was suppressed by siRNA targeted SPRR3 (Figure 4C). Furthermore, the cell invasion was also restored after overexpression of EZH2 in SPRR3 knockdown group (Figure 4D and E). These results suggest that EZH2 acts as a functional downstream of SPRR3.
SPRR3 Is a Downstream Target of miR-876-3p
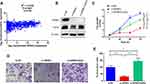
To investigate the upstream of SPRR3, TargetScan was used to predict the targeting miRNAs. Among these miRNAs, miR-876-3p was shown to target SPRR3’s 3ʹUTR with high context score (94) and 8 mer seed match (Figure 5A). As seen in Figure 5B, the dual-luciferase activity was reduced after co-transfection with miR-876-3p. miR-876-3p suppress the wildtype (WT) 3ʹUTR of SPRR3 but not mutant (MUT) 3ʹUTR, suggesting a binding role of miR-876-3p and SPRR3-3ʹUTR. To further confirm the specific post-transcriptional regulation of SPRR3 by miR-876-3p, the protein level of SPRR3 and EZH2 were detected by Western blot. In Figure 5C, miR-876-3p inhibits SPRR3 protein level, indicating SPRR3 is a direct target of miR-876-3p.
miR-876-3p Regulates Cell Proliferation and Invasion of NSCLC
miR-876-3p has been identified as a tumor suppressor in glioma and pancreatic adenocarcinoma.15,16 But the exact role involving in NSCLC is largely unknown. At last, we measured the function of miR-876-3p in NSCLC. As shown in Figure 6A, miR-876-3p was transfected into NSCLC cells and the expression level was up-regulated. As a result, miR-876-3p significantly suppresses cell growth in A549 and PC9 cells (Figure 6B and C). Consistent with this, overexpression of miR-876-3p also inhibits cell invasion (Figure 6D and E), suggesting a tumor suppressor role of miR-876-3p in NSCLC.
Discussion
Carcinogenesis is a complicated process affected by genetic mutation and tumor microenvironment (TME) change in which they are embedded. Accumulated evidence has pointed out that TME factors play an important role in cancer metastasis.17 As we previously described, SPRR3, was screened out and found to be involved in NSCLC cell metastasis and survival.4
A lot of studies demonstrated that SPRR3 supports several types of cancer. Overexpress of SPRR3 was known to promote breast cancer cell lines survival and metastasis, while reduced SPRR3 showed an obviously reduction of tumor xenograft size.18,19 An interesting contradiction is that overexpression of SPRR3 promoting apoptosis in esophageal cancer,5 while in NSCLC, colorectal and breast cancer, elevated expression of SPRR3 is found to be correlated with tumor growth and knockdown of SPRR3 suppresses cell proliferation and invasion, which may as a result of promoted cell apoptosis.7,8 This controversy may be caused by different types of epithelial cells, like squamous and adenocarcinoma, and need further confirmation. Moreover, according to the tumor dormancy that proliferation and metastasis are always uncoupled,20 indicating the invasiveness by knock-down of SPRR3 may not be a result of decreased cell proliferation.
Upon the Uniprot database (https://www.uniprot.org/uniprot/Q9UBC9), SPRR3 is found located in extracellular region and plasma membrane, the mechanism between its expression and cytoplasm function is largely unknown. Several studies indicated that SPRR3 promotes phosphorylation of Akt,7,8 which in turn modulated EZH2 expression, an oncogene in several types of cancer.13 Moreover, inhibition of EZH2 could restrict NSCLC cell proliferation which is a good target in the pharmaceutical industry.14,21-24 These evidences suggest that SPRR3 may promote phosphorylation of Akt and then regulate EZH2 expression.
In addition, by using the online prediction, luciferase-reporter assay and Western blot, we found that SPRR3 is a direct target of miR-876-3p since its 3ʹUTR has binding site (Figure 5A). Several studies suggest that miRNAs play indispensable roles in carcinogenesis and metastasis. In consistent with some other reports that miR-876-3p as a tumor suppressor in glioma and pancreatic adenocarcinoma.15,16 We further validated that miR-876-3p was critical for NSCLC cell growth and invasion (Figure 6). In particular, miR-876-3p could regulate JAG2, a ligand in Notch pathway, which plays important roles in lung cancer.25 These data indicated that miR-876-3p regulates several genes, including SPRR3, and suppresses tumor growth in NSCLC.
Taken together, SPRR3 is overexpressed in NSCLC patients. Further, SPRR3 modulates cell growth and invasion mainly via regulating EZH2. In addition, SPRR3 is predicted and confirmed to be a target of miR-876-3p, a tumor suppressor in NSCLC. This research highlights the potential therapeutic value of miR-876-3p/SPRR3/EZH2 in malignant NSCLC.
Availability of Data
The data that support the findings of this study are available from the corresponding author upon reasonable request. Please contact corresponding author Prof. Yang, if you want to request the dataset.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81972825), Suzhou administration of Science & Technology (SS201762, SYSD20180076), Suzhou Technology development program (ZXL2018171) and the Medical and health technology project of Suzhou New District (2018Q013).
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
RH is supported by GenePharma Co., Ltd. The authors report no other conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Xue X, Fei X, Hou W, Zhang Y, Liu L, Hu R. miR-342-3p suppresses cell proliferation and migration by targeting AGR2 in non-small cell lung cancer. Cancer Lett. 2018;412:170–178. doi:10.1016/j.canlet.2017.10.024
3. Wysoczynski M, Ratajczak MZ. Lung cancer secreted microvesicles: underappreciated modulators of microenvironment in expanding tumors. Int J Cancer. 2009;125(7):1595–1603. doi:10.1002/ijc.v125:7
4. Hu R, Huffman KE, Chu M, Zhang Y, Minna JD, Yu Y. Quantitative secretomic analysis identifies extracellular protein factors that modulate the metastatic phenotype of non-small cell lung cancer. J Proteome Res. 2016;15(2):477–486. doi:10.1021/acs.jproteome.5b00819
5. Zhang Y, Feng YB, Shen XM, et al. Exogenous expression of Esophagin/SPRR3 attenuates the tumorigenicity of esophageal squamous cell carcinoma cells via promoting apoptosis. Int J Cancer. 2008;122(2):260–266. doi:10.1002/ijc.23104
6. Liu Q, Zhang C, Ma G, Zhang Q. Expression of SPRR3 is associated with tumor cell proliferation and invasion in glioblastoma multiforme. Oncol Lett. 2014;7(2):427–432. doi:10.3892/ol.2013.1736
7. Kim JC, Yu JH, Cho YK, et al. Expression of SPRR3 is associated with tumor cell proliferation in less advanced stages of breast cancer. Breast Cancer Res Treat. 2012;133(3):909–916. doi:10.1007/s10549-011-1868-5
8. Cho DH, Jo YK, Roh SA, et al. Upregulation of SPRR3 promotes colorectal tumorigenesis. Mol Med. 2010;16(7–8):271–277. doi:10.2119/molmed.2009.00187
9. Hu X, Peng N, Qi F, Li J, Shi L, Chen R. Cigarette smoke upregulates SPRR3 by favoring c-Jun/Fra1 heterodimerization in human bronchial epithelial cells. Future Oncol. 2018;14(25):2599–2613. doi:10.2217/fon-2018-0043
10. Pyle AL, Atkinson JB, Pozzi A, et al. Regulation of the atheroma-enriched protein, SPRR3, in vascular smooth muscle cells through cyclic strain is dependent on integrin alpha1beta1/collagen interaction. Am J Pathol. 2008;173(5):1577–1588.
11. Huang H, Wang Y, Li Q, Fei X, Ma H, Hu R. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett. 2019;446:81–89. doi:10.1016/j.canlet.2019.01.007
12. Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi:10.1016/j.ccr.2006.01.025
13. Riquelme E, Behrens C, Lin HY, et al. Modulation of EZH2 expression by MEK-ERK or PI3K-AKT signaling in lung cancer is dictated by different KRAS oncogene mutations. Cancer Res. 2016;76(3):675–685. doi:10.1158/0008-5472.CAN-15-1141
14. Zhang H, Qi J, Reyes JM, et al. Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Discov. 2016;6(9):1006–1021. doi:10.1158/2159-8290.CD-16-0164
15. Tang J, Xu J, Zhi Z, et al. MiR-876-3p targets KIF20A to block JAK2/STAT3 pathway in glioma. Am J Transl Res. 2019;11(8):4957–4966.
16. Yang F, Zhao WJ, Jia CL, et al. MicroRNA-876-3p functions as a tumor suppressor gene and correlates with cell metastasis in pancreatic adenocarcinoma via targeting JAG2. Am J Cancer Res. 2018;8(4):636–649.
17. Zhai W, Hu GH, Zheng JH, et al. High expression of the secreted protein dickkopf homolog 4: roles in invasion and metastasis of renal cell carcinoma and its association with Von Hippel-Lindau gene. Int J Mol Med. 2014;33(5):1319–1326. doi:10.3892/ijmm.2014.1673
18. Zweitzig DR, Smirnov DA, Connelly MC, Terstappen LW, O’Hara SM, Moran E. Physiological stress induces the metastasis marker AGR2 in breast cancer cells. Mol Cell Biochem. 2007;306(1–2):255–260.
19. Wang Z, Hao Y, Lowe AW. The adenocarcinoma-associated antigen, AGR2, promotes tumor growth, cell migration, and cellular transformation. Cancer Res. 2008;68(2):492–497. doi:10.1158/0008-5472.CAN-07-2930
20. Yang X, Liang X, Zheng M, Tang Y. Cellular phenotype plasticity in cancer dormancy and metastasis. Front Oncol. 2018;8:505. doi:10.3389/fonc.2018.00505
21. Han Li C, Chen Y. Targeting EZH2 for cancer therapy: progress and perspective. Curr Protein Pept Sci. 2015;16(6):559–570. doi:10.2174/1389203716666150409100233
22. Serresi M, Siteur B, Hulsman D, et al. Ezh2 inhibition in Kras-driven lung cancer amplifies inflammation and associated vulnerabilities. J Exp Med. 2018;215(12):3115–3135. doi:10.1084/jem.20180801
23. Lu X, He X, Su J, et al. EZH2-Mediated epigenetic suppression of GDF15 predicts a poor prognosis and regulates cell proliferation in non-small-cell lung cancer. Mol Ther Nucleic Acids. 2018;12:309–318. doi:10.1016/j.omtn.2018.05.016
24. Geng J, Li X, Zhou Z, Wu CL, Dai M, Bai X. EZH2 promotes tumor progression via regulating VEGF-A/AKT signaling in non-small cell lung cancer. Cancer Lett. 2015;359(2):275–287. doi:10.1016/j.canlet.2015.01.031
25. Yang Y, Ahn YH, Gibbons DL, et al. The notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121(4):1373–1385. doi:10.1172/JCI42579
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.