Back to Journals » Cancer Management and Research » Volume 11
Dysregulation of ncRNAs located at the DLK1-DIO3 imprinted domain: involvement in urological cancers
Authors Li J, Shen H, Xie H, Ying Y , Jin K, Yan H , Wang S, Xu M, Wang X, Xu X, Xie L
Received 13 October 2018
Accepted for publication 6 December 2018
Published 15 January 2019 Volume 2019:11 Pages 777—787
DOI https://doi.org/10.2147/CMAR.S190764
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Chien-Feng Li
Jiangfeng Li,* Haixiang Shen,* Haiyun Xie,* Yufan Ying, Ke Jin, Huaqing Yan, Song Wang, Mingjie Xu, Xiao Wang, Xin Xu, Liping Xie
Department of Urology, First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang Province, China
*These authors contributed equally to this work
Abstract: Genomic imprinting has been found to be involved in human physical development and several diseases. The DLK1-DIO3 imprinted domain is located on human chromosome 14 and contains paternally expressed protein-coding genes (DLK1, RTL1, DIO3) and numerous maternally expressed ncRNA genes (MEG3, MEG8, antisense RTL1, miRNAs, piRNAs, and snoRNAs). Emerging evidence has implicated that dysregulation of the DLK1-DIO3 imprinted domain especially the imprinted ncRNAs is critical for tumor progressions. Multiple miRNAs and lncRNAs have been investigated in urological cancers, of which several are transcribed from this domain. In this review, we present current data about the associated miRNAs, lncRNAs, and piRNAs and the regulation of differentially methylated regions methylation status in the progression of urological cancers and preliminarily propose certain concepts about the potential regulatory networks involved in DLK1-DIO3 imprinted domain.
Keywords: ncRNAs, DLK1-DIO3 imprinted domain, epigenetics, regulatory network, urological cancers, DMRs, MEG3
Introduction
Genomic imprinting usually plays a critical role in human development and diseases. As one of the important imprinted domains, the DLK1-DIO3 imprinted domain is located on human chromosome 14, which contains three paternally expressed protein-coding genes (DLK1, RTL1, and DIO3), maternally expressed lncRNAs (MEG3, MEG8, and antisense RTL1) genes, and numerous sncRNA genes. Emerging evidence implicates this domain in disease pathogenesis, especially in cancers.1,2
ncRNAs refer to RNAs that do not encode proteins but modulate the whole transcription and translation process in an epigenetic way. Currently, 98% DNA in human genome is nonprotein coding, which was once considered as “junk” DNA for a time, with the continuous deeper investigation of ncRNAs, the ENCODE project in 2012 discovered that at least 80% of the human genome is biologically active.3 According to the length of RNAs, they are classified as lncRNAs (>200 bp), and sncRNAs (<200 bp), which include miRNAs, piRNAs, and snoRNAs, and recently, a kind of nonlinear ncRNAs has been detected and described as circular RNAs.4,5
Numerous studies have revealed that the differential distributions and significant roles of ncRNAs are involved in various diseases, especially in cancers, ncRNAs as biomarkers present important diagnostic values. As the largest miRNAs cluster, DLK1-DIO3 imprinted domain is composed of 54 miRNAs, 11 lincRNAs, and several piRNAs and snoRNAs, and surging evidence has indicated that deregulation of these ncRNAs in this region is significantly associated with tumor progression.1,6 Similarly, an amount of ncRNAs especially miRNAs is identified to be involved in urological cancers; besides, these miRNAs are partially transcribed from DLK1-DIO3 imprinted domain.
In this review, we provide an overview of the representative ncRNAs at DLK1-DIO3 imprinted domain and the regulation networks involved in urological cancers (RCC, BCa, and PCa).
DLK1-DIO3 imprinted domain
Genomic imprinting and ncRNAs at DLK1-DIO3 imprinted domain
Genomic imprinting
Genomic imprinting is a critical epigenetics-regulating phenomenon that contributes to specific, monoallelic gene expression in diploid cells.7 It has been a consensus that imprinted genes have profound effects on fetal development, placental biology, and controlling of the activities in neonates, such as the feeding, maintenance of body temperature, and regulation of metabolism. In addition, its regulatory values are gradually demonstrated in a wide range of common diseases, especially in obesity, diabetes mellitus, cancer, etc.8 Nearly 150 imprinted genes have been identified in the mouse (data in MouseBook Imprinting Catalog), and ~50% of these genes have nearly also been detected in humans (data in Catalog of Parent of Origin Effects).9 Furthermore, >80% of the known imprinted genes are characteristic of cluster, and about 2–15 genes which always contain two or more genes, and the size of each cluster vary from <100 kb to several megabases.10 DLK1-DIO3 imprinted domain is one of the clustered genomic imprinting regions, which is located on distal mouse chromosome 12 and human chromosome 14.11 Its critical regulatory role in physical development and several diseases are being uncovered step by step.
ncRNAs at DLK1-DIO3 imprinted domain
Currently, the paternally expressed genes DLK1, RTL1, and DIO3, and the maternally expressed ncRNAs MEG3 (Gtl2), MEG8 (RIAN), and antisense RTL1 (RTL1as) are detected in this domain. Moreover, numerous sncRNAs also enrich the contents of this domain. So far, there are 53 miRNAs on the forward strand and one more (miR-1247) on the reverse strand, which consist of the largest human miRNAs cluster, another nine lncRNAs, of which six are on the forward strand (CTD-2561F5.1, RP11-168L7.3, RP11-909M7.2, AL132709.5, AL132709.7, and AL132709.8) and three on the reverse strand (AL132709.1, RP11-168L7.1, and DIO3OS), several snoRNAs downstream of MEG3, piRNAs, and several pseudogenes, which are all encoded at the DLK1-DIO3 domain (Figure 1).
  | Figure 1 The schematic diagram of DLK-DIO3 imprinted domain including multiple ncRNAs and upstream DMRs. Abbreviation: DMR, differentially methylated region. |
Regulations of DLK1-DIO3 imprinted domain
Allele-specific expression of imprinted domain is mainly regulated by ICRs, most of which are DMRs in the DLK1-DIO3 imprinted region. According to their origin, DMRs are classified as the gDMRs including IG-DMR, H19-ICR, KvDMR1, Nespas-DMR, and somatic DMRs including GTL2-DMR, and DLK1-DMR.12,13 Three methylated DMRs: the IG-DMR, DLK1-DMR, and the MEG3-DMR are detected to regulate the DLK1-DIO3 imprinted region in humans. Several critical regulatory proteins, histone modifications, and lncRNAs/miRNAs participate in the regulation of DMRs methylation.
Several critical regulatory proteins
Several critical regulatory proteins are involved in the process of genomic imprinting. DNA methyltransferases (DNMT3A/B) are dispensable for the establishment step with their catalytic functions. Besides, certain indirect regulatory proteins are also crucial for the maintenance of this domain. As a KRAB-contained zinc finger protein, ZFP57 is vital to maintain the parent-of-origin-specific epigenetic marks and expression of the imprinted gene clusters by selectively binding to the methylated motif (TGC5mCGC).14–17 Another key protein KAP1 is reported to selectively maintain the stability of DMRs that are effective in H19-ICR and H19 promoter DMR but do not function in IG-DMR and Peg3-DMR.18 Multiple ways may exist for PGC7 in the regulation of DMRs. Previous study has indicated that PGC7 recognizes and binds H3K9me2 and corresponding chromatins to protect the methylation of the imprinted domains.19 But recently, its N-terminal DNA-binding domain has been detected to interact with TET2 and TET3 to suppresses the enzymatic activity of these two proteins, thus partially protecting the methylation status of DMRs.20 The regulation mediated by the PRC2 is still controversial. Conventional theory proposes that PRC2 serves as a maintenance of the repressor to control domain silencing via catalyzing H3K2me3, and a further RIP sequencing assay confirms that the 9,000 lncRNAs are involved in this regulation (such as Xist, H19, Igf2, MEG3); thus, at DIO3-DLK1 imprinted domain, PRC2 can repress DLK1 expression by interacting with MEG3 lncRNAs.21,22 However, a recent study provides a novel mechanism of PRC2 in the regulation of the maternal Gtl2-Rian-Mirg locus, suggesting that PRC2 maintains expression of the locus by counteracting DNMT3 to prevent their recruitments and subsequent DNA methylation at the IG-DMR,23 which indicates the complicated regulations of DLK1-DIO3 imprinting domain. In terms of the erasure of imprinted domain, previous findings detect the partial erasing role of 5-methylcytosine oxidizing TET1 enzyme at some imprinted regions.24–27 However, silencing of TET1 does not appear to upregulate the methylation status at DLK1-DIO3 DMR, but significantly upregulate the methylation status of H19 DMR in uES, dES and EB cell lines.28
Histone modifications
Histone methylation is usually interacted with DNA methylation.29 Substantial methylations of H3K4 induced by the histone lysine demethylase KDM1B deficiency block the formation of DMR methylations during oogenesis.30 In addition, variable histone modifications were detected in paternally and maternally methylated ICRs during the stages preceding the global histone-to-protamine exchange. H3-lysine-4 methylation and H3 acetylation are abundant at maternally methylated ICRs, but absent at paternally methylated ICRs.31
Regulatory role of lncRNAs/miRNAs
A recent in vitro experiment reveals that ~30 variable genes including DLK1, DIO3, and MEG3 are detected upregulated in HOTAIR-targeted deletion mice, and silencing of HOTAIR disables the binding to the PRC2 and LSD1 complex, consequently causing H3K4me3 gain and H3K27me3 loss at Dlk1 imprinted gene to upregulate its expression; however, no significant alterations of the IG-DMR methylation status are detected.32 Interestingly, both the miR-127 and the miR-136 of this domain are detected to target RTL1, which reveals that self-regulation mediated by miRNAs transcribed from this domain also exist. In addition, PRC2 (RBAP48, EED, and HDAC2) are found targeted by miR-495 and miR-323-3p of this domain, presenting a feedback regulatory loop in regulating all the genes and ncRNAs encoded by this DLK1-DIO3 domain in full pluripotent stem cells.
Deregulations of DLK1-DIO3 imprinted domain are involved in various diseases especially cancers
DLK1-DIO3 imprinted domain and diseases
It has been established that DLK1-DIO3 imprinted domain is critical not only for the normal growth but also the cognitions and behaviors after birth. The dysregulations of this domain are associated with several rare cognitive disorders and even common diseases ranging from obesity, diabetes mellitus, psychiatric disorders, and cancers.2,8,33 In terms of cancers, dysregulations of ncRNAs in this domain, especially miRNAs and lncRNAs, the paternally expressed genes DLK1, RTL1, and DIO3, and the domain regulatory factors such as the modification enzymes and key proteins (such as KAP1) are all involved in the tumorigenesis of cancers, which indicates a complicated regulation network in tumor progression.
Mechanisms of the DLK1-DIO3 imprinted domain in regulating cancers
Reviewing the previous studies, we found that disordered DMR regulations are the concentrated mechanisms to dysregulate the expression of certain mRNA or ncRNA expression, subsequently inducing cancers. Although many studies found that the imprinted genes are associated with cancers, however, mechanisms by which imprinting is regulated in cancers are poorly understood. Apart from the above complicated regulations of DMRs from variable approaches that are involved in carcinogenesis, other points are listed as follows.
Methylation status of DMRs
Previous studies have confirmed that the hypermethylation status of DMRs at the DLK1-DIO3 imprinted domain significantly reduced the expression of certain tumor suppressor miRNA clusters in several cancers.34–37 It has been proven that MEG3 and DLK1 are deregulated in urothelial cancer due to the epigenetic silencing including hypermethylation of IG-DMRs and DLK1 promoter and repressive histone modifications across the 14q32 imprinted gene cluster.38 Similarly, in BCa, hypermethylated status of IG-DMR is also found to be an inhibitor for the expression of miR-323a-3p that repressed the EMT progression.
LOI
LOI induced by abnormal methylation status or mutations of DMRs has been described to contribute to the carcinogenesis and cancer progression.39–41 LOI is identified as an important risk factor for colorectal carcinogenesis, and similarly, LOI of different loci has been confirmed to promote the progression in oligodendrogliomas, breast cancer, and hepatocellular carcinomas.42–45 Upregulated IGF2 induced by LOI has been reported to be significantly associated with few tumors such as the rhabdomyosarcoma, adrenal cancer, colon cancer and PCa, squamous cell carcinoma of the head and neck.46–50 Because of the structural similarity between IGF2 and DLK1, the LOI of DMR is also consistently detected at the DLK1-GTL2 locus in embryonal rhabdomyosarcomas but erasure of imprinting in alveolar rhabdomyosarcoma, which leads to a higher GTL2/DLK1 mRNA ratio in alveolar rhabdomyosarcoma.46 And LOI at the DLK1-MEG3 domain generates the widespread epigenetic instability through deregulated expression in human hepatocellular carcinoma.51 In addition, genome-wide miRNA profiles suggest that LOI changes in the 14q32 noncoding region are helpful to define and classify the osteosarcoma subtypes; however, the precise regulation network relationship needs further investigations.52
Dysregulation of ncRNAs at the DLK1-DIO3 imprinted domain contributes to the carcinogenesis of urological cancers
As previous findings describe, numerous miRNAs are distributed in cluster at DLK1-DIO3 imprinted domain. Besides, dysregulation of these miRNA clusters due to abnormal DMR methylations is involved in several cancers including urological cancers (RCC, BCa, and PCa). ncRNAs composed of lncRNAs, miRNAs, piRNAs, and snoRNAs are the important epigenetic regulators in multiple cancers. So far, the largest miRNAs cluster is detected in this domain. In this section, we will review the dysregulated ncRNAs of this cluster at the DLK1-DIO3 imprinting domain in urological cancers (Table 1) (Figure 2).
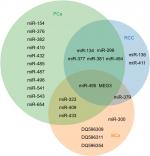  | Figure 2 The common or specific ncRNAs in each class of urological cancers. Abbreviations: BCa, bladder cancer; PCa, prostate cancer; RCC, renal cell carcinoma. |
RCC
Dysregulations of several ncRNAs have been confirmed to be associated with RCC, especially the disordered miRNAs of the DLK1-DIO3 imprinted domain. In this section, we review the associated ncRNAs of this domain in regulating RCC.
Previously, certain studies have found that miR-136 is downregulated in ccRCC, which suggests to be a novel biomarker. Overexpression of miR-136 significantly induced apoptosis and inhibited the proliferation and migration.53–55 Similarly, Kaplan–Meier survival analysis reveals that upregulated miR-379 predicts a poor overall survival in pRCC.56 A significantly repressed proliferation, migration, and induced apoptosis is observed in RCC cell (786-O and ACHN) with miR-411 mimics.57 Eleven miRNAs are screened and identified to be associated with the tumor progression from ccRCC clinical tumor samples, among which miR-299-3 p is obviously downregulated.58 Forced expression of miR-494 inhibits the survival of RCC cell accompanied by increased lipid droplets and mitochondrial changes.59 The tumor suppressing effect of miR-495 on proliferation in RCC has been confirmed and can be repressed by lncRNA UCA1 via RNA sponging effect.60 miR-381 has been proven to be a WEE inhibitor suppressing the proliferation and inducing the sensitivity to 5-FU by upregulating CDC2 level in RCC cell line (786-O).61,62 It has been reported that miR-134 inhibits the cell proliferation and EMT progression by directly targeting KRAS.63 miR-377 was significantly downregulated in ccRCC samples and gain of function further validates that miR-377 significantly inhibits the proliferation and migration by targeting E26 transformation-specific-1.64 Apart from the regulation of various miRNAs in this region, lncRNAs also play a great role. Previous research has reported that 2.5% Wilms’ tumor samples present a hypermethylation status in MEG3-DMR, which significantly inhibits the expression of tumor-suppressed MEG3, thus triggering the tumor progression.65 Further study demonstrate that inhibition of MEG3 repressed the targeted imprinted genes (IGF2 in Wilms tumor and DLK1 in RCC), and DLK1 is lost in 78% primary RCC tissues and identified as a critical tumor suppressor in the regulation of RCC.65 A consistent function assay confirms that overexpression of MEG3 induces the cell apoptosis via mitochondrial pathway in 786-0 cell lines. To conclude, MEG3 plays a crucial tumor suppressor role in RCC.
Bladder cancer
Previously, it has been proven that 14q loss is common in invasive BCa, and two potential suppressor loci at 14q12 and 14q32.1–32.2 are detected.66 Numerous ncRNAs especially miRNAs have been reported to be involved in tumorigenesis of BCa, several of which belong to DLK1-DIO3 region. The associated ncRNAs of this domain in regulating BCa are reviewed in this section.
Forced expression of miR-409 is found to inhibit the EMT progression by directly targeting c-MET.67 Hypermethylation of miR-433 promoter has been confirmed to contribute to the downregulation of miR-433 in BCa, and further experiment reveals that both c-MET and CREB1 are the direct targets of miR-433; besides, MITF is detected as an intermediate protein induced by CREB1 to indirectly regulate c-MET/AKT/GSK-3β/SNAIL signaling, thus a complicated network mediated by miR-433 is established to regulate the proliferation and EMT progression of BCa.68 Interestingly, miR-323a-3p in this region is also identified to regulate the EMT progression by targeting c-MET and SMAD3. In addition, SNAIL is the last confocal protein found to induce EMT, thus, miR-323a-3p/c-MET/SMAD3/SNAIL circuit is detected.37 We find that miR-409, miR-433, miR-323a-3p all target c-MET to inhibit EMT of BCa. In addition, overexpression or silencing of c-MET represses or induces the expression of above miRNAs in turn. However, its specific mechanisms still remain elusive, and a critical network may be involved. The tumor-suppressive role of miR-379-5p in tumor proliferation and metastasis is demonstrated to target MDM2, and overexpression of MDM2 partially reverses the inhibitory effects of miR-379-5p, which further validates the direct interaction between miR-379-5p and MDM2.69 A recent study analyses the 67 pairs of BCa and adjacent normal bladder tissues to find miR-495 as an onco-miRNA that is upregulated in BCa tissues and overexpression of miR-495 promotes BCa progression via targeting PTEN, which is significantly negatively correlated with miR-495.70 By utilizing miRNA microarray, researchers detected the expression pattern of miRNAs in normal tissues (5 samples) and BCa (30 samples), and found that miR-300 is downregulated in all six groups (grade I, grade II, grade III, grade I+II+III, infiltrating, and noninfiltrating group) and further confirmed by q-RT-PCR in BCa cell line T24.71 Similar to RCC, MEG3 also plays a tumor suppressor role in BCa progression. Previous function study proves that downregulation of MEG3 induces autophagy but contributes to cell proliferation in BCa.72 A recent lncRNAs doublecheck screening between 80 BCa with matched adjacent normal tissues, and 220 BCa patients’ serum samples, suggests that the expression of MEG3 is negatively correlated with recurrence-free survival by Kaplan–Meier analysis, which proposes that MEG3 is recurrence-independent prognostic factor.73 In terms of the mechanisms of dysregulation of MEG3, both hypermethylation of IG-DMRs and DLK1 promoter and repressive histone modifications contribute to the loss or downregulation of MEG3 and DLK1. Accumulating evidence has confirmed that piRNAs inhibit the target gene transcription or translation in cancer progression via targeting DNA sequence with piRNA/PIWI complex or targeting 3′-UTR with RNA-induced silencing complex. One hundred thirty-eight piRNAs are detected at the DLK1-DIO3 imprinted domain. Recent study reports that seven piRNAs are differentially expressed and predict the patient outcome of lung cancer combined with several miRNAs in this domain. However, currently, no study refers to piRNAs of this domain in BCa progression.74 However, a set of BCa piRNA microarray data have been reported, by analyzing the data, we found that three piRNAs transcribed from the DLK-DIO3 domain, DQ596309 (fold change =17.2, P=0.017), DQ596354 (fold change =6.2, P=0.027), and DQ596311 (fold change =5.3, P=0.031) are downregulated in BCa,74 which needs a further investigation of function and mechanisms. To conclude, piRNAs in this domain may serve as critical potential tumor regulators in BCa tumorigenicity.
Prostate cancer
In this section, we conduct a whole review of ncRNAs in the regulation of PCa. Previous study has conducted a systematic screening about the miRNA expression at 14q32.31 in four cell lines (normal prostate epithelial cells, three PCa cell lines DU-145, PC3, and LNCaP), and the results found that almost all miRNAs in this region are downregulated except for miR-656 in PC3 cell line.75,76 In addition, after the treatment of AZA and the histone deacetylase inhibitor trichostatin, an obvious elevation of above miRNA cluster is detected, which indicates a potential epigenetic silencing results in the deregulations of miRNAs in this region. Further analysis of the expression pattern of these miRNAs (miR-154, miR-299-5 p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3 p, miR-495, and miR-654-3 p) correlates with clinical and pathological variables.76 Circulating miR-433 and miR-495 from PCa serum samples present their capability in differentiating indolent (low-risk) and aggressive (high-risk) PCa, and the results suggest that in high-risk PCa, circulating miR-433 is upregulated but miR-495 is downregulated.77 In addition, another study reports that AKT and mTOR can be downregulated by miR-495 to inhibit the growth and migration.78 miR-432 upregulated by LncRNA625 has been found to inhibit PCa cell proliferation by targeting TRIM29 and PYGO2 to regulate Wnt/β-catenin pathway.79 Elevated miR-154-3 p and miR-379 is observed to promote PCa bone metastasis, and its expression level is significantly negatively correlated with progression-free survival of PCa patients.80 However, several studies have confirmed that miR-154 inhibits proliferation via targeting CCND2 and inhibits EMT progression via HMGA2.81,82 E2F5 can also be downregulated by miR-154 to reduce the proliferation and migration,82 which present the multitargets of miRNAs in the regulation of PCa. miR-323a is found to be upregulated in PCa tissues and overexpression of miR-323 increases the VEGF-A level and the vessel formation by targeting adiponectin receptor.83 Downregulation of CXCR4 mediated by miR-494 significantly represses the PCa growth and metastasis, which may be a potential therapeutic target for the treatment of PCa.84 Synergistic effects are obtained after cotransfecting miR-494 (survivin targeted) with survivin short hairpin RNA to inhibit the growth of PC3 cell line.85 miRNAs in this domain are also associated with androgen bioavailability. UGT is defined as a critical modulation factor to control androgen bioavailability, and interestingly, miR-409 and miR-494 are detected to target UGT2B17 UGT and miR-376c target both UGT2B15 and UGT2B17.86 Besides, ectopic expression of miR-409 induced by stromal fibroblast promotes the tumor induction and EMT.87 miR-543 has been identified a EMT promotor in PCa LNCAP and C4-2B cell lines via directly downregulating Raf kinase inhibitory protein.88 Downregulation of SPAG5 induced by miR-539 drastically blocked PCa progressions, which maybe a therapeutic target.89 miR-382 also shows its tumor suppressor role in cell proliferation and metastasis by targeting the chicken ovalbumin upstream promoter transcription factor II.90 miR-134, an RAS-targeted miRNA, has been reported to be repressed with other several miRNAs by inorganic arsenic during the transformation from the human prostate epithelial line to CAsE-PE and from the derivative normal stem cell line to As-cancer stem cell line.91 Recently, a meta-analysis of PCa miRNA expression profiles suggest that 22 miRNAs including miR-496 and miR-541 are downregulated in recurrent PCa samples compared with nonrecurrent PCa samples, which may provide new candidate biomarkers for predicating recurrent PCa.92 A study enrolling 149 PCa patients and 178 controls indicates that circulating miR-410 is drastically upregulated in PCa patients and may serve as a specific diagnostic biomarker of PCa (area under curve of 0.8097 (95% CI, 0.7371–0.8823; P<0.001).93 Recently, miR-493-5p is found to be dysregulated by the hypermethylation of its promotor, and ectopic expression of miR-493-5p targets c-MET, CREB1, and EGFR to inhibit cell proliferation and migration via downregulating AKT/GSK-3β/SNAIL signaling.94 Interestingly, MEG3 is also involved in the progression of PCa. Previous analysis of 14 microarray has indicated that MEG3 combined with eight other imprinted genes are downregulated, which was further validated by q-RT-PCR assay.95,96 A function study demonstrates that forced expression of MEG3 significantly induces G0/G1 phase arrest and apoptosis of PCa cell.97 One of the critical mechanism of LncRNA is RNA sponge effect that functions as a ceRNA. And numerous imbalanced ceRNA pairs are found to be involved in the progression of PCa, among which loss of MEG3-AQP3 ceRNA pair is identified as having potential prognostic value.98
Conclusion
To conclude, accumulating evidence has implicated the significant role of ncRNAs at the DLK1-DIO3 imprinted domain in the regulation of urological cancers tumorigenicity. From the above review, we find that miRNAs and lncRNAs are the dominant regulators among all regulatory factors, while studies on the piRNAs and snoRNAs in urological cancers are still few. Besides, the regulatory mechanisms are especially complicated, which indicates that the potential networks may exist in the whole progression and needs further investigations.
Throughout the review, we propose that several individual concepts or problems should be solved. 1) We review three common cancers of urology; however, the functions of the same miRNA may differ in different cancers. For example, miR-409 is a tumor suppressor in BCa, but it promotes the EMT progression in PCa, which indicate the tumor specificity and the regulation complexities of this domain. 2) In terms of the dysregulation of ncRNAs in this region, we find that hypermethylation of DMRs mainly accounts for this phenomenon of urological cancers. It is the same condition of the normal physiological process that DMRs are hypermethylated after paternal transmission and hypomethylated after maternal transmission in somatic cells. In addition, IG-DMR appears at the control of methylation pattern of the imprinting regulator of both paternally and maternally expressed genes, ie, of the secondary MEG3-DMR on the maternally transmitted chromosome.99,100 Recently, MEF2A was found to induce the expression of miRNAs in DLK-DIO3 involved in skeletal muscle regeneration via binding to MEG3 promoter.101,102 The molecular signaling network may contribute to carcinogenesis and tumor progression. We find that miR-409, miR-433, and miR-323a-3p own the common target c-MET, and c-MET can also negatively modulate the expression of these miRNAs in BCa. Consistently, c-MET is also one of the targets of miR-493-5 p; however, whether c-MET can similarly regulate miR-493-3 p has yet to be elucidated. Interestingly, Kumar et al identify a negative feedback loop between miR-493 and IGF1R via SNAIL in head and neck cancers, which means that miR-493 can target IGF1R/AKT/GSK-3β/NF-KB/SNAIL to inhibit SNAIL on the one hand, while on the other hand, SNAIL can bind to the promoter of miR-493 to inhibit the expression of miR-493 in turn.103 Concerning that miR-493, miR-409, miR-433, and miR-323a-3p are located at the close positions of DLK1-DIO3 imprinted domain, which gives us a hint as to whether c-MET can consistently inhibit the expression of miR-409/miR-433/miR-323a-3p by inducing SNAIL to bind to the promoter region of them. Thus, miRNAs/c-MET/SNAIL negative feedback loop maybe involved in the DLK1-DIO3 imprinted domain of urological cancers; however, above concepts may need more studies to demonstrate. 3) We find most of the miRNAs in this region of PCa show downregulation, which indicates the tumor suppressor role. We wonder if these are potential therapeutic targets that can be activated by certain relatively mature genomic technique. 4) A large number of miRNAs have been detected; however, more concentrations need to be attached to the studies of snoRNAs and piRNAs, which may play a great role in the regulation of the whole imprinted region. 5) Although numerous ncRNAs and their functions have been confirmed, it is still a puzzle as to how the expression pattern and the epigenetic regulations are involved in urological cancers. No doubt DLK1-DIO3 presents a significant effect on the progressions of cancers including urological cancer, but it is still a long way to explore the profound mechanism and clinical therapeutic values in the future.
Abbreviations
AZA, 5-aza-2′-deoxycytidine; BCa, bladder cancer; ccRCC, clear cell renal cell carcinoma; ceRNA, competing endogenous RNAs; CAsE-PE, chronic arsenic-exposed prostate epithelial; DMR, differentially methylated region; EMT, epithelial–mesenchymal transition; ETS1, E26 transformation specific-1; gDMRs, germline DMRs; IG-DMR, intergenic DMR; ICR, imprinting control region; IGF2, insulin-like growth factor 2; LOI, loss of imprinting; lncRNAs, long noncoding RNAs; miRNAs, microRNAs; ncRNAs, noncoding RNAs; PCa, prostate cancer; piRNA, PIWI-interacting RNA; PRC2, polycomb repressive complex 2; pRCC, papillary renal cell carcinoma; RCC, renal cell carcinoma; RIP, RNA immunoprecipitation; sncRNA, small noncoding RNA; snoRNAs, small nucleolar RNAs; UGT, UDP-glucuronosyltransferases conjugating enzymes.
Acknowledgments
This review was supported by grants from the National Natural Science Foundation of China (81702500, 81772744), Zhejiang Province Medical and Health Scientific Research Project (2018KY032), and the Zhejiang Province Key Project of Science and Technology (2014C04008-2).
Disclosure
The authors report no conflicts of interest in this work.
References
Benetatos L, Hatzimichael E, Londin E, et al. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70(5):795–814. | ||
Enterina JR, Enfield KSS, Anderson C, Marshall EA, Ng KW, Lam WL. DLK1-DIO3 imprinted locus deregulation in development, respiratory disease, and cancer. Expert Rev Respir Med. 2017;11(9):749–761. | ||
Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337(6099):1159–1161. | ||
Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38(5):485–491. | ||
Zheng Y, Liu L, Shukla GC. A comprehensive review of web-based non-coding RNA resources for cancer research. Cancer Lett. 2017;407:1–8. | ||
Enfield KS, Martinez VD, Marshall EA, et al. Deregulation of small non-coding RNAs at the DLK1-DIO3 imprinted locus predicts lung cancer patient outcome. Oncotarget. 2016;7(49):80957–80966. | ||
Konyukhov BV, Platonov ES. Genomic imprinting in mammals. Genetika. 1992;18(1):203. | ||
Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15(8):517–530. | ||
Weksberg R. Imprinted genes and human disease. Am J Med Genet C Semin Med Genet. 2010;154C(3):317–320. | ||
Barlow DP. Genomic imprinting: a mammalian epigenetic discovery model. Annu Rev Genet. 2011;45(1):379–403. | ||
da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24(6):306–316. | ||
Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19(3):281–289. | ||
Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12(8):565–575. | ||
Quenneville S, Verde G, Corsinotti A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44(3):361–372. | ||
Zuo X, Sheng J, Lau HT, et al. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem. 2012;287(3):2107–2118. | ||
Li X, Ito M, Zhou F, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15(4):547–557. | ||
Strogantsev R, Krueger F, Yamazawa K, et al. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 2015;16(1):112. | ||
Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335(6075):1499–1502. | ||
Nakamura T, Liu YJ, Nakashima H, et al. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486(7403):415–419. | ||
Bian C, Yu X. PGC7 suppresses TET3 for protecting DNA methylation. Nucleic Acids Res. 2014;42(5):2893–2905. | ||
Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. | ||
Zhao J, Ohsumi TK, Kung JT, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. | ||
das PP, Hendrix DA, Apostolou E, et al. PRC2 is required to maintain expression of the maternal Gtl2-Rian-Mirg locus by preventing de novo DNA methylation in mouse embryonic stem cells. Cell Rep. 2015;12(9):1456–1470. | ||
Dawlaty MM, Breiling A, Le T, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24(3):310–323. | ||
Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504(7480):460–464. | ||
Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263(1):6–21. | ||
Hackett JA, Sengupta R, Zylicz JJ, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339(6118):448–452. | ||
Liu L, Mao SQ, Ray C, et al. Differential regulation of genomic imprinting by TET proteins in embryonic stem cells. Stem Cell Res. 2015;15(2):435–443. | ||
Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta. 1839;2014(12):1362. | ||
Ciccone DN, Su H, Hevi S, et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461(7262):415–418. | ||
Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 2007;26(3):720–729. | ||
Li L, Liu B, Wapinski OL, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5(1):3–12. | ||
Brandvain Y, van Cleve J, Ubeda F, Wilkins JF. Demography, kinship, and the evolving theory of genomic imprinting. Trends Genet. 2011;27(7):251–257. | ||
Lehner B, Kunz P, Saehr H, Fellenberg J. Epigenetic silencing of genes and microRNAs within the imprinted Dlk1-Dio3 region at human chromosome 14.32 in giant cell tumor of bone. BMC Cancer. 2014;14(1):495. | ||
Berenstein R, Blau O, Nogai A, et al. Multiple myeloma cells alter the senescence phenotype of bone marrow mesenchymal stromal cells under participation of the DLK1-DIO3 genomic region. BMC Cancer. 2015;15(1):68. | ||
An F, Yamanaka S, Allen S, et al. Silencing of miR-370 in human cholangiocarcinoma by allelic loss and interleukin-6 induced maternal to paternal epigenotype switch. PLoS One. 2012;7(10):e45606. | ||
Li J, Xu X, Meng S, et al. MET/SMAD3/SNAIL circuit mediated by miR-323a-3p is involved in regulating epithelial-mesenchymal transition progression in bladder cancer. Cell Death Dis. 2017;8(8):e3010. | ||
Greife A, Knievel J, Ribarska T, Niegisch G, Schulz WA. Concomitant downregulation of the imprinted genes DLK1 and MEG3 at 14q32.2 by epigenetic mechanisms in urothelial carcinoma. Clin Epigenetics. 2014;6(1):29–13. | ||
Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. Relaxation of imprinted genes in human cancer. Nature. 1993;362(6422):747–749. | ||
Sakatani T, Kaneda A, Iacobuzio-Donahue CA, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307(5717):1976–1978. | ||
Steshina EY, Carr MS, Glick EA, Yevtodiyenko A, Appelbe OK, Schmidt JV. Loss of imprinting at the Dlk1-Gtl2 locus caused by insertional mutagenesis in the Gtl2 5’ region. BMC Genet. 2006;7(1):44. | ||
Sakatani T, Kaneda A, Iacobuziodonahue CA. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2011;64(2):208–214. | ||
de Souza AT, Yamada T, Mills JJ, Jirtle RL. Imprinted genes in liver carcinogenesis. FASEB J. 1997;11(1):60–67. | ||
Kobatake T, Yano M, Toyooka S, et al. Aberrant methylation of p57KIP2 gene in lung and breast cancers and malignant mesotheliomas. Oncol Rep. 2004;12(5):1087–1092. | ||
Hsu CM, Lin PM, Lin HC, et al. Altered expression of imprinted genes in squamous cell carcinoma of the head and neck. Anticancer Res. 2016;36(5):2251–2258. | ||
Schneider G, Bowser MJ, Shin DM, Barr FG, Ratajczak MZ. The paternally imprinted DLK1-GTL2 locus is differentially methylated in embryonal and alveolar rhabdomyosarcomas. Int J Oncol. 2014;44(1):295–300. | ||
Nielsen HM, How-Kit A, Guerin C, et al. Copy number variations alter methylation and parallel IGF2 overexpression in adrenal tumors. Endocr Relat Cancer. 2015;22(6):953–967. | ||
Nishihara S, Hayashida T, Mitsuya K, et al. Multipoint imprinting analysis in sporadic colorectal cancers with and without microsatellite instability. Int J Oncol. 2000;17(2):317–322. | ||
Wang H, Ge S, Qian G, et al. Restoration of IGF2 imprinting by polycomb repressive complex 2 docking factor SUZ12 in colon cancer cells. Exp Cell Res. 2015;338(2):214–221. | ||
Damaschke N, Yang B, Bhusari S. Loss of Igf2 gene imprinting in murine prostate promotes widespread neoplastic growth. Cancer Res. 2017;77(19):5236–5247. | ||
Anwar SL, Krech T, Hasemeier B, et al. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS One. 2012;7(11):e49462. | ||
Hill KE, Kelly AD, Kuijjer ML, et al. An imprinted non-coding genomic cluster at 14q32 defines clinically relevant molecular subtypes in osteosarcoma across multiple independent datasets. J Hematol Oncol. 2017;10(1):107. | ||
Liu H, Brannon AR, Reddy AR, et al. Identifying mRNA targets of microRNA dysregulated in cancer: with application to clear cell renal cell carcinoma. BMC Syst Biol. 2010;4(1):51. | ||
Xing T, He H. Epigenomics of clear cell renal cell carcinoma: mechanisms and potential use in molecular pathology. Chin J Cancer Res. 2016;28(1):80. | ||
Chen P, Zhao L, Pan X. Tumor suppressor microRNA–136–5p regulates the cellular function of renal cell carcinoma. Oncol Lett. 2018;15(4):5995–6002. | ||
Ge YZ, Xu LW, Xu Z, et al. Expression profiles and clinical significance of microRNAs in papillary renal cell carcinoma: a STROBE-compliant observational study. Medicine. 2015;94(16):e767. | ||
Zhang X, Zhang M, Cheng J, Lv Z, Wang F, Cai Z. MiR-411 functions as a tumor suppressor in renal cell cancer. Int J Biol Markers. 2017;32(4):e454–e460. | ||
He H, Wang L, Zhou W, et al. MicroRNA expression profiling in clear cell renal cell carcinoma: identification and functional validation of key miRNAs. PLoS One. 2015;10(5):e0125672. | ||
Dutta P, Haller E, Sharp A, Nanjundan M. MIR494 reduces renal cancer cell survival coinciding with increased lipid droplets and mitochondrial changes. BMC Cancer. 2016;16(1):1–14. | ||
Lu Y, Liu W-G, Lu J-H, et al. LncRNA UCA1 promotes renal cell carcinoma proliferation through epigenetically repressing p21 expression and negatively regulating miR-495. Tumor Biol. 2017;39(5):101042831770163. | ||
Chen B, Duan L, Yin G, Tan J, Jiang X. Simultaneously expressed miR-424 and miR-381 synergistically suppress the proliferation and survival of renal cancer cells – Cdc2 activity is up-regulated by targeting WEE1. Clinics. 2013;68(6):825–833. | ||
Chen B, Duan L, Yin G, Tan J, Jiang X. miR-381, a novel intrinsic WEE1 inhibitor, sensitizes renal cancer cells to 5-FU by up-regulation of Cdc2 activities in 786-O. J Chemother. 2013;25(4):229–238. | ||
Liu Y, Zhang M, Qian J, et al. miR-134 functions as a tumor suppressor in cell proliferation and epithelial-to-mesenchymal transition by targeting KRAS in renal cell carcinoma cells. DNA Cell Biol. 2015;34(6):429–436. | ||
Wang R, Ma Y, Yu D, Zhao J, Ma P. miR-377 functions as a tumor suppressor in human clear cell renal cell carcinoma by targeting ETS1. Biomed Pharmacother. 2015;70:64–71. | ||
Astuti D, Latif F, Wagner K, et al. Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. Br J Cancer. 2005;92(8):1574–1580. | ||
Chang WY, Cairns P, Schoenberg MP, Polascik TJ, Sidransky D. Novel suppressor loci on chromosome 14q in primary bladder cancer. Cancer Res. 1995;55(15):3246–3249. | ||
Xu X, Chen H, Lin Y, et al. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells. 2013;36(1):62–68. | ||
Xu X, Zhu Y, Liang Z, et al. c-Met and CREB1 are involved in miR-433-mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3β/Snail signaling. Cell Death Dis. 2016;7(2):e2088. | ||
Wu D, Niu X, Tao J, et al. MicroRNA-379-5p plays a tumor-suppressive role in human bladder cancer growth and metastasis by directly targeting MDM2. Oncol Rep. 2017;37(6):3502–3508. | ||
Tan M, Mu X, Liu Z, et al. microRNA-495 promotes bladder cancer cell growth and invasion by targeting phosphatase and tensin homolog. Biochem Biophys Res Commun. 2017;483(2):867–873. | ||
Xie P, Xu F, Cheng W, et al. Detection, verification and significance of differentially expressed miRNAs in bladder urothelial carcinoma. Zhonghua Yi Xue Za Zhi. 2010;90(48):3391–3394. | ||
Ying L, Huang Y, Chen H, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9(3):407. | ||
Duan W, du L, Jiang X, et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2016;7(48):78850–78858. | ||
Chu H, Hui G, Yuan L, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356(2):561–567. | ||
Formosa A, Lena AM, Markert EK, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32(1):127–134. | ||
Formosa A, Markert EK, Lena AM, et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33(44):5173–5182. | ||
Alhasan AH, Scott AW, Wu JJ, et al. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc Natl Acad Sci USA. 2016;113(38):10655–10660. | ||
Li JZ, Wang ZL, Xu WH, Li Q, Gao L, Wang ZM. MicroRNA-495 regulates migration and invasion in prostate cancer cells via targeting Akt and mTOR signaling. Cancer Invest. 2016;34(4):181. | ||
Li JB, Liu F, Zhang BP, et al. LncRNA625 modulates prostate cancer cells proliferation and apoptosis through regulating the Wnt/β-catenin pathway by targeting miR-432. Eur Rev Med Pharmacol Sci. 2017;21(11):2586–2595. | ||
Gururajan M, Josson S, Chu GC, et al. miR-154* and miR-379 in the DLK1-DIO3 microRNA mega-cluster regulate epithelial to mesenchymal transition and bone metastasis of prostate cancer. Clin Cancer Res. 2014;20(24):6559–6569. | ||
Zhu C, Shao P, Bao M, et al. miR-154 inhibits prostate cancer cell proliferation by targeting CCND2. Urol Oncol. 2014;32(1):31.e9-16. | ||
Zhu C, Li J, Cheng G, et al. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem. 2013;379(1–2):69–75. | ||
Gao Q, Yao X, Zheng J. MiR-323 inhibits prostate cancer vascularization through adiponectin receptor. Cell Physiol Biochem. 2015;36(4):1491–1498. | ||
Shen PF, Chen XQ, Liao YC, et al. MicroRNA-494-3p targets CXCR4 to suppress the proliferation, invasion, and migration of prostate cancer. Prostate. 2014;74(7):756–767. | ||
Zhu J, Sun C, Wang L, et al. Targeting survivin using a combination of miR494 and survivin shRNA has synergistic effects on the suppression of prostate cancer growth. Mol Med Rep. 2016;13(2):1602–1610. | ||
Margaillan G, Lévesque É, Guillemette C, Guillaume M, Éric L, Chantal G. Epigenetic regulation of steroid inactivating UDP-glucuronosyltransferases by microRNAs in prostate cancer. J Steroid Biochem Mol Biol. 2016;155(Pt A):85–93. | ||
Josson S, Gururajan M, Sung SY, et al. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene. 2015;34(21):2690–2699. | ||
du Y, Liu XH, Zhu HC, Wang L, Ning JZ, Xiao CC. MiR-543 promotes proliferation and epithelial-mesenchymal transition in prostate cancer via targeting RKIP. Cell Physiol Biochem. 2017;41(3):1135–1146. | ||
Zhang H, Li S, Yang X, Qiao B, Zhang Z, Xu Y. miR-539 inhibits prostate cancer progression by directly targeting SPAG5. J Exp Clin Cancer Res. 2016;35(1):60. | ||
Zhang W, Liu J, Qiu J, et al. MicroRNA-382 inhibits prostate cancer cell proliferation and metastasis through targeting COUP-TFII. Oncol Rep. 2016;36(6):3707–3715. | ||
Ngalame NN, Tokar EJ, Person RJ, Xu Y, Waalkes MP. Aberrant microRNA expression likely controls RAS oncogene activation during malignant transformation of human prostate epithelial and stem cells by arsenic. Toxicol Sci. 2014;138(2):268–277. | ||
Pashaei E, Pashaei E, Ahmady M, Ozen M, Aydin N. Meta-analysis of miRNA expression profiles for prostate cancer recurrence following radical prostatectomy. PLoS One. 2017;12(6):e0179543. | ||
Wang J, Ye H, Zhang D, et al. MicroRNA-410-5p as a potential serum biomarker for the diagnosis of prostate cancer. Cancer Cell Int. 2016;16(1):1–6. | ||
Wang S, Wang X, Li J. c-Met, CREB1 and EGFR are involved in miR-493-5p inhibition of EMT via AKT/GSK-3β/Snail signaling in prostate cancer. Oncotarget. 2017;8(47):82303–82313. | ||
Ribarska T, Bastian KM, Koch A, Schulz WA. Specific changes in the expression of imprinted genes in prostate cancer--implications for cancer progression and epigenetic regulation. Asian J Androl. 2012;14(3):436–450. | ||
Ribarska T, Goering W, Droop J, Bastian KM, Ingenwerth M, Schulz WA. Deregulation of an imprinted gene network in prostate cancer. Epigenetics. 2014;9(5):704–717. | ||
Luo G, Wang M, Wu X, et al. Long non-coding RNA MEG3 inhibits cell proliferation and induces apoptosis in prostate cancer. Cell Physiol Biochem. 2015;37(6):2209–2220. | ||
Liu D, Yu X, Wang S, et al. The gain and loss of long noncoding RNA associated-competing endogenous RNAs in prostate cancer. Oncotarget. 2016;7(35):57228. | ||
Kagami M, O’Sullivan MJ, Green AJ, et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 2010;6(6):e1000992. | ||
Benetatos L, Hatzimichael E, Londin E, et al. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70(5):795–814. | ||
Snyder CM, Rice AL, Estrella NL, Held A, Kandarian SC, Naya FJ. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development. 2013;140(1):31–42. | ||
Wüst S, Dröse S, Heidler J, et al. Metabolic maturation during muscle stem cell differentiation is achieved by miR-1/133a-mediated inhibition of the Dlk1-Dio3 mega gene cluster. Cell Metab. 2018;27(5):1026–1039. | ||
Kumar AS, Jagadeeshan S, Pitani RS, et al. Snail-modulated MicroRNA 493 forms a negative feedback loop with the insulin-like growth factor 1 receptor pathway and blocks tumorigenesis. Mol Cell Biol. 2017;37(6):e00510-16. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.