Back to Journals » Journal of Asthma and Allergy » Volume 14
Dynamic Urinary Proteome Changes in Ovalbumin-Induced Asthma Mouse Model Using Data-Independent Acquisition Proteomics
Authors Qin W, Wang T, Liu G, Sun L, Han W, Gao Y
Received 18 July 2021
Accepted for publication 30 October 2021
Published 9 November 2021 Volume 2021:14 Pages 1355—1366
DOI https://doi.org/10.2147/JAA.S330054
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Amrita Dosanjh
Weiwei Qin,1,2 Ting Wang,2 Guangwei Liu,3 Lixin Sun,1 Wei Han,4 Youhe Gao2
1Department of Anesthesiology, Qingdao Municipal Hospital, Qingdao University, Qingdao, 266071, People’s Republic of China; 2Department of Biochemistry and Molecular Biology, Gene Engineering Drug and Biotechnology Beijing Key Laboratory, Beijing Normal University, Beijing, 100875, People’s Republic of China; 3Key Laboratory of Cell Proliferation and Regulation Biology, Ministry of Education, Institute of Cell Biology, College of Life Sciences, Beijing Normal University, Beijing, 100875, People’s Republic of China; 4Department of Respiratory Medicine, Qingdao Municipal Hospital, Qingdao University, Qingdao, 266071, People’s Republic of China
Correspondence: Youhe Gao; Wei Han Tel +86 10 58804382
; +86 532 85937809
Email [email protected]; [email protected]
Background: In this work, we aim to investigate dynamic urinary proteome changes during asthma development and to identify potential urinary protein biomarkers for the diagnosis of asthma.
Methods: An ovalbumin (OVA)-induced mouse model was used to mimic asthma. The urinary proteome from asthma and control mice was determined using data-independent acquisition combined with high-resolution tandem mass spectrometry.
Results: Overall, 331 proteins were identified, among which 53 were differentially expressed (26, 24, 14 and 20 on days 2, 8, 15 and 18, respectively; 1.5-fold change, adjust P< 0.05). Gene Ontology annotation of the differential proteins showed that the acute-phase response, innate immune response, B cell receptor signaling pathway, and complement activation were significantly enriched. Protein–protein interaction network revealed that these differential proteins were partially biologically connected in OVA-induced asthma, as a group. On days 2 and 8, after two episodes of OVA sensitization, six differential proteins (CRAMP, ECP, HP, F2, AGP1, and CFB) were also reported to be closely associated with asthma. These proteins may hold the potential for the early screening of asthma. On days 15 and 18, after challenged with 1% OVA by inhalation, seven differential proteins (VDBP, HP, CTSE, PIGR, AAT, TRFE, and HPX) were also reported to be closely associated with asthma. Thus, these proteins hold the potential to be biomarkers for the diagnosis of asthma attack.
Conclusion: Our results indicate that the urinary proteome could reflect dynamic pathophysiological changes in asthma progression.
Keywords: OVA-induced asthma, mice, urine, proteome, data-independent acquisition
Introduction
Asthma is a heterogeneous disease that is usually characterized by chronic airway inflammation. It is characterized by variable symptoms, such as wheeze, dyspnea, and chest tightness and/or cough that vary over time and intensity, together with variable expiratory airflow limitation.1 Asthma is a major public health concern worldwide, with global prevalence ranging from 1% to 21% in adults2 and up to 32% in children aged 6–7 years.3 The predominant mechanism involved in the pathogenesis of asthma is a Type 2 helper T cell (Th2) cytokine-mediated eosinophilic airway inflammation associated with hyper-responsiveness of the lungs.4 Asthma is a heterogeneous disease; many clinical phenotypes of asthma have been identified, including allergic and nonallergic asthma, late-onset asthma, asthma with persistent airflow limitation, and asthma with obesity.5,6 Many of these and other phenotypes of asthma are diagnosed based on the clinical characteristics.
Spirometry is by far the primary diagnostic approach for asthma, which reflects disease severity rather than disease activity.7 Moreover, its measurements strictly depend on patient’s compliance, physician’s expertise, and data interpretation.8 Especially for children aged under 6 years, the diagnosis is more difficult because they are too young to undergo spirometry. Besides, clinical symptoms on which pulmonologists rely for a correct diagnostic approach are subjective and nonspecific. The differential diagnosis in a patient with suspected asthma is based on the age of onset, reversibility of airflow limitation, symptom variability, and atopy. A significant proportion of patients with chronic respiratory symptoms (9–55%) has diagnoses and/or features of both asthma and chronic obstructive pulmonary disease (COPD).9 Distinguishing asthma from chronic airflow limitation due to COPD becomes problematic, particularly among smokers.10,11 Thus, there is an urgent need for noninvasive biomarkers for early diagnosis, evaluation of lung functional impairment, and prognosis in asthma.
Mass spectrometry (MS)-based proteomics has dramatically improved and emerged as a prominent tool in the field of biomarker study. Many protein biomarkers of asthma and/or COPD have been described and categorized as primarily blood and bronchoalveolar lavage fluid (BALF) biomarkers.12,13 For example, MS–based proteomic analysis revealed serum ApoE to be significantly downregulated in atopic asthmatics compared to healthy volunteers.14 Comparative proteomics analysis of patients with quick development and slow development COPD, provided 15 potential serum biomarkers for efficiently predicting the development phenotypes of COPD.15
Urine, as an attractive resource for biomarker research, can be collected noninvasively and continuously, which has arrested more and more concerns. Mass spectrometry-based proteomics based on the association between different protein profiles and pathogenic processes is gaining an important role in pulmonary medicine.16–18 Urinary proteomic studies have identified several candidate biomarkers for pulmonary disease, such as lung cancer,19 pulmonary fibrosis, and tuberculosis.20,21 However, several candidate biomarkers of asthma have been described primarily in the sputum, blood, and bronchoalveolar lavage fluid. Besides, the urinary proteome could reflect changes in disease progression at the early stage. For example, the urinary proteome profiles changed significantly even in the absence of clinical manifestations or histopathological damage to the lung tissue in bleomycin-induced pulmonary fibrosis21 and Walker-256 tail-vein injection-induced lung metastasis22 rat models. Therefore, urine can sensitively reflect pathophysiological changes in the lung tissue at an early stage and is, thus, a promising resource for studying the biomarkers of lung diseases.
This study aimed to observe dynamic urinary proteome changes during asthma development and to identify potential urinary protein biomarkers for the diagnosis of asthma. The data-independent acquisition (DIA) approach was used to profile the urinary proteome of OVA-induced asthma mouse model.
Materials and Methods
Mice and Asthma Induction
The C57BL/6J male mice (6-week-old) were purchased from Charles River (Beijing, China) and housed in light (12 h light: 12 h dark cycle) and temperature-controlled (22 ± 3 °C) rooms. Throughout the experiments, the mice had free access to standard laboratory chow and were provided with tap water ad libitum. The mice were treated humanely according to international guidelines and the experiment was adhered to the Guide for the Care and Use of Laboratory Animals and approved by the Animal Ethics Committee of Qingdao Municipal Hospital, Qingdao, China.
Twelve specific pathogen-free C57BL/6 mice were used and divided randomly into control (n=6) and asthma (n=6) groups. During the induction of asthma, the mice were sensitized by intraperitoneal injection of 50 µg ovalbumin grade V (OVA) (Sigma, Germany) adsorbed on Al(OH)3 (Alumlnject©, Perbio, Belgium) on days 1 and 7. Animals were fed 50 µg of OVA via nasal drops on days 14–17, as previously described.23 Each control mouse was administered 0.9% NaCl as placebo.
Histological Analysis
On day 17, the mice were anesthetized by intraperitoneal injection of 10% pentobarbital. After anesthesia induction, a catheter was intubated through the trachea. The left lung was removed after ligation of the left main bronchus, and the lung tissues were then quickly fixed in 10% neutral-buffered formalin. The formalin-fixed tissues were embedded in paraffin sectioned (4 mm) and stained with hematoxylin and eosin (H&E) to identify histopathological lesions.
ELISA and Flow Cytometry
The blood samples were collected by cardiac puncture, and centrifuged at 3000 rpm for 5 min at 4 °C. IgE level was measured by an ELISA kit (BD Bioscience) according to the manufacturer’s instructions.
After the left lung was removed, 1 mL of iced PBS was slowly injected into the trachea with a syringe. The bronchoalveolar lavage fluid (BALF) was washed several times, counted on a blood cell counting plate, and 5×105 cells were added to the 7AAD-percP (BD Biosciences), CD45-eflour 450 (Biolegend), CD11b-ITC (Biolegend), and SiglecF-PE (Biolegend), according to the specification. Antibody surface staining of BALF cells. After 30 min of light staining on ice, 200 µL PBS was added, the solution was centrifugated at 5000 r/min for 5 min, and excess antibody was washed off. The cells were resuspended in 300 μL of PBS or a 1:1 mixture of PBS and 4% paraformaldehyde and then transferred to a flow tube for on-machine detection using flow cytometry (ACEA Novo CyteTM).
Urine Collection and Sample Preparation
Urine samples were collected from six asthmatic mice at days 2, 8, 15, and 18 and six control mice. The mice were individually housed in the metabolic cages for 4 hours. During urine collection, no food was provided to the mice to avoid urine contamination. After collection, the urine samples were centrifuged at 2000 g for 30 min at 4°C immediately and then stored at −80°C.
For tryptic digestion, the proteins were digested with trypsin (Promega, USA) using filter-aided sample preparation methods.24 Briefly, 100 µg of the protein sample was loaded on the 10-kD filter unit (Pall, USA). The protein solution was reduced with 4.5 mM DTT for 1 h at 37°C and then alkylated with 10 mM of indoleacetic acid for 30 min at room temperature in the dark. The proteins were digested with trypsin (enzyme-to-protein ratio of 1:50) for 14 h at 37°C. The peptides were desalted on Oasis HLB cartridges (Waters, USA) and lyophilized for trap column fractionation and LC-DIA-MS/MS analysis.
Spin Column Separation
The pooled peptides sample were fractionated using a high-pH reversed-phase peptide fractionation kit (Thermo Pierce, USA) according to the manufacturer’s instructions. Briefly, 60 µg of the pooled peptide sample was loaded onto the spin column. Ten different fractions were collected by centrifugation, including flow-through fraction, wash fraction, and eight step gradient sample fractions (5, 7.5, 10, 12.5, 15, 17.5, 20, and 50% acetonitrile). The fractionated samples were lyophilized and used for the LC-DDA-MS/MS analysis.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
The Orbitrap Fusion Lumos Tribrid Mass Spectrometer coupled with an EASY-nLC 1200 HPLC system (Thermo Scientific, Germany) was used for the DDA-MS and DIA-MS analysis. The digested peptides were dissolved in 0.1% formic acid and loaded on a trap column (75 µm × 2 cm, 3 µm, C18, 100 A°). The eluent was transferred to a reversed-phase analytical column (50 µm × 250 mm, 2 µm, C18, 100 A°). The eluted gradient was 5–30% buffer B for 90 min. The calibration kit (iRT kit from Biognosys, Switzerland) was spiked at a concentration of 1:20 v/v in all samples. DDA-MS: full scan, 350–1550 m/z at 60,000; cycle time: 3 secs (top speed mode); AGC 2e5; MS/MS scans in the Orbitrap at a resolution of 15,000; isolation window 2 Da, HCD 32%, AGC 5e5, 50 ms. DIA-MS: 32 windows.
DIA Quantification Analysis
The raw data files were processed using Proteome Discoverer (version 2.1; Thermo Scientific, Germany) with SEQUEST HT against the SwissProt ratus database (released in May 2019, containing 8086 sequences). The search parameters consisted of the parent ion mass tolerance, 10 ppm; fragment ion mass tolerance, 0.02 Da; fixed modifications, carbamidomethylated cysteine (+58.00 Da); and variable modifications, oxidized methionine (+15.995 Da) and deamidated glutamine and asparagine (+0.984 Da). Other settings included the default parameters.25 The DIA-MS raw files were imported to Spectronaut Pulsar with the default settings. In brief, all results were filtered by an FDR of 1%. Peptide intensity was calculated by summing the peak areas of their respective fragment ions for MS2.
Bioinformatics Analysis
Bioinformatics analysis was conducted in order to better study the biological function of the dysregulated proteins. Gene Ontology (GO) analysis was performed on the urinary differentially altered proteins identified at the discovery phase (http://www.geneontology.org/).26,27 Protein-protein interaction (PPI) networks were constructed using the STRING database (http://www.string-db.org), which is a database of known and predicted protein interactions, including direct (physical) and indirect (functional) associations.
The “Wu Kong” platform (https://www.omicsolution.org/wkomics/main/) was used for statistical analysis. The differential proteins were selected using one-way ANOVA, and p-values were adjusted using the Benjamini–Hochberg method. Significance was set at a p-value of < 0.05 and a fold change of 1.5.
Results
Characterization of OVA-Induced Asthmatic Mice
To investigate the dynamic urinary proteome changes in asthma progression, we established an OVA-induced asthma model. After challenging with OVA, the serum IgE levels were measured (Figure 1A). The results showed that the IgE levels in the OVA-treated group were increased. Next, flow cytometry was performed on eosinophils in BALF. After removing the dead cells (7AAD+), the proportion of eosinophils in the leukocytes (CD45+) was analyzed (Figure 1B). The ratio of eosinophils (CD11b+siglecF+) in BLAF was 67.75% after 17 days of OVA induction.
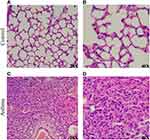
The hematoxylin-eosin (HE) stained microscopic features of the lung from control mice showed a normal structure: the alveolar wall structure is intact, there is no thickening, and there is no exudate in the alveolar cavity (Figure 2A), and alveolar epithelial cells are arranged in order (Figure 2B). Compared to the control group, HE-stained of the lung in asthma group showed alveolar destruction and alveolar wall thickening (Figure 2C), as well as pulmonary fibrosis and neutrophil infiltration (Figure 2D). These pathological changes reveal the success of OVA-induced asthma modeling.
Dynamic Urinary Proteome Changes in OVA-Induced Asthmatic Mice
For a preliminary investigation of changes in the urinary proteome with asthma progression, three urine samples from the control group and 12 samples taken at four time points (days 2, 8, 15, and 18) from the asthma group were analyzed by LC-DIA-MS/MS workflow.
Overall, a total of 331 urinary protein groups were identified; all details are listed in Supporting Table S1. Among these, 53 proteins were significantly changed in asthmatic mice compared to control mice (1.5-fold change, adjust P<0.05; Table 1).
 |
Table 1 Details of the Differential Urinary Proteins in OVA-Induced Asthmatic Mice |
At day 2, 26 differential proteins, 21 of which increased and five of which decreased, were identified. At day 8, 24 differential proteins, 15 of which increased and nine of which decreased, were identified. At day 15, 14 differential proteins, six of which increased and nine of which decreased, were identified. At day 18, 20 differential proteins, seven of which increased and 13 of which decreased, were identified (Figure 3).
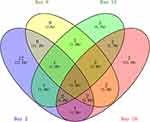 |
Figure 3 Vein diagram of the differential urinary proteins in asthmatic mice compared with control mice. |
Gene Ontology Analysis of Differential Proteins
The GO functional annotation was performed on the differentially expressed proteins in asthmatic mice. A total of 53 dysregulated proteins, at four time points, were annotated and classified to be involved with certain biological processes (Figure 4).
 |
Figure 4 GO analysis of the differential proteins at days 2, 8, 15 and 18 in OVA-induced mice. (A) Biological process; (B) Cellular component; (C) Molecular function. |
The GO enrichment analysis showed that inflammation or immune responses, such as acute-phase response, innate immune response, B cell receptor signaling pathway, complement activation, positive regulation of B cell activation, and phagocytosis, were the mainly involved biological processes at the four time points. Coagulation factor II, haptoglobin, and alpha-1-acid glycoprotein 1 were enriched in acute-phase response. Beta-2 microglobulin, cathelicidin antimicrobial peptide, complement factor B, immunoglobulin heavy chain, immunoglobulin heavy constant gamma 2A, immunoglobulin kappa constant, and lipocalin 2 were enriched in innate immune response. These results indicate that urine can reflect biological responses in the body during the progression of asthma.
In the cellular component category, most of these differential proteins were extracellular region, extracellular space, blood microparticle, extracellular exosome, and immunoglobulin complex proteins (Figure 4B). In the molecular function category, serine-type endopeptidase activity, immunoglobulin receptor binding, hormone activity, and serine-type peptidase activity were overrepresented on days 2 and 8. Immunoglobulin receptor binding, insulin-activated receptor activity, small molecule binding, and transporter activity were overrepresented on days 15 and 18 (Figure 4C).
Protein-Protein Interaction Network of the Differential Proteins
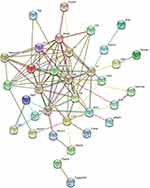
To better understand the pathogenic mechanisms of OVA-induced asthma, the PPI network for 53 differential proteins was constructed using STRING (Figure 5). The STRING PPI network analysis revealed that the average node degree is 3.84, average local clustering coefficient is 0.443, and PPI enrichment p-value is <1.0e-16. This indicates that these proteins have more interactions among themselves than what would be expected for a random set of proteins of similar size drawn from the genome.
Differential Urinary Proteins Related to Asthma
Among these 53 differential proteins, 18 that had human orthologs were reported to be closely associated with asthma (Table 2). The first 13 differential proteins shown in the table were changed on days 2 and 8, which was the early stage at which a low dose of OVA was intraperitoneally injected for sensitization of mice, and the last seven differential proteins were changed on days 15 and 18, during which a high dose of OVA was administered via nasal inhalation to provoke asthma.
 |
Table 2 Differential Urinary Proteins Related to Asthma |
Discussion
In this study, urine samples were collected from six control mice and six asthmatic mice on days 2, 8, 15, and 18 for proteome analysis to identify potential biomarkers for asthma. Compared to control mice, 53 proteins were found to have undergone significant changes in the urine samples of asthmatic mice.
On days 2 and 8, 13 differential proteins that had human orthologs were reported to be closely associated with asthma. Among these, six urinary proteins were changed on days 2 and 7, and the trend was consistent at each time point; these proteins hold the potential to be biomarkers for the early screening of asthma attack. For example, cathelicidin antimicrobial peptide (CRAMP) significantly enhanced the infiltration of inflammatory cells and accumulation of pro-inflammatory Th2 cytokine IL-13 and IL-33 in BALF and exacerbated lung tissue inflammation and airway goblet cell hyperplasia in a previous study.28 CRAMP plays an important role in the accumulation of inflammatory monocyte-derived dendritic cells in the peribronchiolar tissue in allergic airway inflammation.29 Serum eosinophil cationic protein 1 (ECP) was positively correlated with the severity of an asthma attack.30,31 Elevated serum ECP level had the highest likelihood of fixed airflow obstruction among patients with asthma.32 Serum ECP level is helpful in assessing the efficacy of inhaled corticosteroids therapy in bronchial asthma.33 Increased serum haptoglobin (HP) level is seen in patients with asthma, asthma exacerbation and 24 hours after allergen challenge in late responders.34 As part of its tissue repair function, HP can induce differentiation of fibroblast progenitor cells into lung fibroblasts in airway remodeling in patients with asthma.35 Patients with asthma had 61% higher maximal prothrombin (PT) conversion rate than controls.36 Plasma PT level was significantly higher in patients with nonallergic asthma than in controls.37 Alpha-1-acid glycoprotein 1 was altered in patients with atopic asthma, and the presence of asthma symptoms correlated with increased glycan branching of AGP in both plasma and BALF.38 Rhinovirus infection is the most common trigger of the acute exacerbations of asthma; the infection stimulated stimulates the expression of not only complement factor B but also the novel pro-inflammatory cytokine IL-32.39
On days 15 and 18, the acute attack stage on which a high-dose of OVA was nasal inhalation to provoke asthma, seven differential proteins that had human orthologs were reported to be closely associated with asthma. Besides HP and vitamin D-binding protein (VDBP), which were altered on day 2, another five differential proteins also showed changes; these proteins hold the potential to be biomarkers for the diagnosis of asthma attack. For example, serum VDBP was significantly upregulated in the steroid-resistant asthma group compared with the steroid-sensitive asthma group in a previous study.40 Another study reported upregulated VDBP expression both in serum and BALF of isocyanate-occupational asthma (OA) patients compared with controls.41,42 These findings were consistent with those of studies in which VDBP was identified in the BALF of patients with asthma using proteomic analysis.43,44 In vivo, Cathepsin E deficiency resulted in reduced lymphocyte influx after allergen sensitization and challenge in both investigated airway inflammation models.45 Polymeric immunoglobulin receptor protein level was decreased in the bronchial epithelium from patients with asthma,46 whereas PIGR demonstrated significantly enhanced upregulation in the BALF of OVA-sensitized mice.47 The prevalence of alpha-1-antitrypsin 1–4 deficiency was low in patients with severe persistent asthma.48 Serotransferrin was upregulated in patients with methylene diphenyl diisocyanate (MDI)-induced occupational asthma and was used as serologic markers of MDI-OA.49 Hemopexin was upregulated in the local lymph nodes and serum of toluene-2,4-diisocyanate-induced asthmatic mice.50 Individuals with asthma–COPD overlap exhibited significantly elevated serum levels of HPX compared with healthy control subjects.51
Urine can be collected noninvasively and continuously, which is a good resource of biomarker for asthma. Despite the advantage of urine as a promising biomarker source, urine proteins are affected by many factors. Therefore, to sort out factors associated with any particular pathophysiological condition, especially in human samples, is challenging. To circumvent this problem, simpler systems, such as animal models, should be used to establish the direct relation between disease progression and urine changes. To avoid contamination, the mice were individually housed in the metabolic cages, and no food was provided during urine collection. To avoid protein degradation, the urine samples were centrifuged immediately after 4 hours collection, and the stored at −80°C.
Overall, this study was a preliminary study with a small number of OVA-induced asthmatic mice. In future studies, the urinary protein biomarkers should be further evaluated in urine samples of patients with asthma to test their sensitivity and specificity. The findings of these studies will have potential applications in monitoring in treatment and prevention studies.
Conclusions
Our results suggest that the urinary proteome could reflect dynamic pathophysiological changes in asthma; therefore, these differential proteins could be potential biomarkers for the early diagnosis of asthma.
Acknowledgments
The National Key Research and Development Program of China (2018YFC0910202, 2016YFC1306300); National Natural Science Foundation of China (82000881); Beijing Natural Science Foundation (7172076); Beijing Cooperative Construction Project (110651103); Beijing Normal University (11100704).
Disclosure
The authors declare that they have no competing interests.
References
1. Bousquet J, Humbert M. GINA 2015: the latest iteration of a magnificent journey. Eur Respir J. 2015;46(3):579–582. doi:10.1183/13993003.01084-2015
2. To T, Stanojevic S, Moores G, et al. Global asthma prevalence in adults: findings from the cross-sectional world health survey. BMC Public Health. 2012;12(1):204. doi:10.1186/1471-2458-12-204
3. Lai CK, Beasley R, Crane J, et al. Global variation in the prevalence and severity of asthma symptoms: phase three of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax. 2009;64(6):476–483. doi:10.1136/thx.2008.106609
4. Abramson MJ, Perret JL, Dharmage SC, McDonald VM, McDonald CF. Distinguishing adult-onset asthma from COPD: a review and a new approach. Int J Chron Obstruct Pulmon Dis. 2014;9:945–962. doi:10.2147/COPD.S46761
5. Moore WC, Meyers DA, Wenzel SE, et al. Identification of asthma phenotypes using cluster analysis in the severe asthma research program. Am J Respir Crit Care Med. 2010;181(4):315–323. doi:10.1164/rccm.200906-0896OC
6. Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18(5):716–725. doi:10.1038/nm.2678
7. Levy ML, Quanjer PH, Booker R, et al.; A General Practice Airways Group (GPIAG)1 document, in association with the Association for Respiratory Technology & Physiology (ARTP)2 and Education for Health. Diagnostic spirometry in primary care: proposed standards for general practice compliant with American Thoracic Society and European Respiratory Society recommendations. Prim Care Respir J. 2009;18(3):130–147.
8. Miller MR, Hankinson J, Brusasco V, et al. Standardisation of spirometry. Eur Respir J. 2005;26(2):319–338. doi:10.1183/09031936.05.00034805
9. Inoue H, Nagase T, Morita S, Yoshida A, Jinnai T, Ichinose M. Prevalence and characteristics of asthma-COPD overlap syndrome identified by a stepwise approach. Int J Chron Obstruct Pulmon Dis. 2017;12:1803–1810. doi:10.2147/COPD.S133859
10. Silva GE, Sherrill DL, Guerra S, Barbee RA. Asthma as a risk factor for COPD in a longitudinal study. Chest. 2004;126(1):59–65. doi:10.1378/chest.126.1.59
11. Zeki AA, Schivo M, Chan A, Albertson TE, Louie S. The asthma-COPD overlap syndrome: a common clinical problem in the elderly. J Allergy. 2011;2011:861926.
12. Serban KA, Pratte KA, Bowler RP. Protein biomarkers for COPD outcomes. Chest. 2021;159(6):2244–2253. doi:10.1016/j.chest.2021.01.004
13. Crameri R. The potential of proteomics and peptidomics for allergy and asthma research. Allergy. 2005;60(10):1227–1237. doi:10.1111/j.1398-9995.2005.00873.x
14. Bhowmik M, Majumdar S, Dasgupta A, Gupta Bhattacharya S, Saha S. Pilot-scale study of human Plasma proteomics identifies ApoE And IL33 as markers in atopic asthma. J Asthma Allergy. 2019;12:273–283. doi:10.2147/JAA.S211569
15. Li F, Xu D, Wang J, Jing J, Li Z, Jin X. Comparative proteomics analysis of patients with quick development and slow development Chronic Obstructive Pulmonary Disease (COPD). Life Sci. 2020;256:117829. doi:10.1016/j.lfs.2020.117829
16. de Torre C, Ying SX, Munson PJ, Meduri GU, Suffredini AF. Proteomic analysis of inflammatory biomarkers in bronchoalveolar lavage. Proteomics. 2006;6(13):3949–3957. doi:10.1002/pmic.200500693
17. Sim SY, Choi YR, Lee JH, et al. In-depth proteomic analysis of human bronchoalveolar lavage fluid toward the biomarker discovery for lung cancers. Proteomics Clin Appl. 2019;13(5):e1900028. doi:10.1002/prca.201900028
18. Norman KC, Moore BB, Arnold KB, O’Dwyer DN. Proteomics: clinical and research applications in respiratory diseases. Respirology. 2018;23(11):993–1003. doi:10.1111/resp.13383
19. Zhang C, Leng W, Sun C, et al. Urine proteome profiling predicts lung cancer from control cases and other tumors. EBioMedicine. 2018;30:120–128. doi:10.1016/j.ebiom.2018.03.009
20. Wang J, Zhu X, Xiong X, et al. Identification of potential urine proteins and microRNA biomarkers for the diagnosis of pulmonary tuberculosis patients. Emerg Microbes Infect. 2018;7(1):63. doi:10.1038/s41426-018-0066-5
21. Wu J, Li X, Zhao M, Huang H, Sun W, Gao Y. Early detection of urinary proteome biomarkers for effective early treatment of pulmonary fibrosis in a rat model. Proteomics Clin Appl. 2017;11(11–12). doi:10.1002/prca.201700103
22. Wei J, Ni N, Meng W, Gao Y. Early urine proteome changes in the Walker-256 tail-vein injection rat model. Sci Rep. 2019;9(1):13804. doi:10.1038/s41598-019-50301-1
23. Kumar RK, Herbert C, Foster PS. The “classical” ovalbumin challenge model of asthma in mice. Curr Drug Targets. 2008;9(6):485–494. doi:10.2174/138945008784533561
24. Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi:10.1038/nmeth.1322
25. He B, Shi J, Wang X, Jiang H, Zhu HJ. Label-free absolute protein quantification with data-independent acquisition. J Proteomics. 2019;200:51–59. doi:10.1016/j.jprot.2019.03.005
26. Consortium TGO. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338.
27. Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi:10.1038/75556
28. Li Y, Chu X, Liu C, et al. Exogenous murine antimicrobial peptide CRAMP significantly exacerbates Ovalbumin-induced airway inflammation but ameliorates oxazolone-induced intestinal colitis in BALB/c mice. Hum Vaccin Immunother. 2018;14(1):146–158. doi:10.1080/21645515.2017.1386823
29. Chen K, Liu M, Liu Y, et al. Signal relay by CC chemokine receptor 2 (CCR2) and formylpeptide receptor 2 (Fpr2) in the recruitment of monocyte-derived dendritic cells in allergic airway inflammation. J Biol Chem. 2013;288(23):16262–16273. doi:10.1074/jbc.M113.450635
30. Mogensen I, Alving K, Bjerg A, et al. Simultaneously elevated exhaled nitric oxide and serum-eosinophil cationic protein relate to recent asthma events in asthmatics in a cross-sectional population-based study. Clin Exp Allergy. 2016;46(12):1540–1548. doi:10.1111/cea.12792
31. Zedan M, Settin A, El-Chennawi F, El-Desouky T, Nasef N, Fouda A. Eosinophilic cationic protein: is it useful in assessing control of childhood asthma? East Mediterr Health J. 2010;16(10):1045–1049. doi:10.26719/2010.16.10.1045
32. Mogensen I, Alving K, Dahlen SE, et al. Fixed airflow obstruction relates to eosinophil activation in asthmatics. Clin Exp Allergy. 2019;49(2):155–162. doi:10.1111/cea.13302
33. Guo CL, Sun XM, Wang XW, Guo Q. Serum eosinophil cationic protein is a useful marker for assessing the efficacy of inhaled corticosteroid therapy in children with bronchial asthma. Tohoku J Exp Med. 2017;242(4):263–271. doi:10.1620/tjem.242.263
34. Lee SH, Kim KH, Kim JM, et al. Relationship between group-specific component protein and the development of asthma. Am J Respir Crit Care Med. 2011;184(5):528–536. doi:10.1164/rccm.201006-0951OC
35. Larsen K, Macleod D, Nihlberg K, et al. Specific haptoglobin expression in bronchoalveolar lavage during differentiation of circulating fibroblast progenitor cells in mild asthma. J Proteome Res. 2006;5(6):1479–1483. doi:10.1021/pr050462h
36. Bazan-Socha S, Mastalerz L, Cybulska A, et al. Asthma is associated with enhanced thrombin formation and impaired fibrinolysis. Clin Exp Allergy. 2016;46(7):932–944. doi:10.1111/cea.12734
37. Asero R, Tedeschi A, Cugno M. Markers of autoreactivity, coagulation and angiogenesis in patients with nonallergic asthma. Allergy. 2011;66(10):1339–1344. doi:10.1111/j.1398-9995.2011.02668.x
38. Van Den Heuvel MM, Poland DC, De Graaff CS, et al. The degree of branching of the glycans of alpha(1)-acid glycoprotein in asthma. A correlation with lung function and inflammatory parameters. Am J Respir Crit Care Med. 2000;6(161):1972–1978.
39. Herbert C, Do K, Chiu V, et al. Allergic environment enhances airway epithelial pro-inflammatory responses to rhinovirus infection. Clin Sci. 2017;131(6):499–509. doi:10.1042/CS20160939
40. Jiang H, Chi X, Zhang X, Wang J. Increased serum VDBP as a risk predictor for steroid resistance in asthma patients. Respir Med. 2016;114:111–116. doi:10.1016/j.rmed.2016.03.011
41. Hur GY, Park HS. Biological and genetic markers in occupational asthma. Curr Allergy Asthma Rep. 2015;15(1):488. doi:10.1007/s11882-014-0488-7
42. Kim SH, Choi GS, Nam YH, et al. Role of vitamin D-binding protein in isocyanate-induced occupational asthma. Exp Mol Med. 2012;44(5):319–329. doi:10.3858/emm.2012.44.5.036
43. Wu J, Kobayashi M, Sousa EA, et al. Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol Cell Proteomics. 2005;4(9):1251–1264. doi:10.1074/mcp.M500041-MCP200
44. Noël-Georis I, Bernard A, Falmagne P, Wattiez R. Database of bronchoalveolar lavage fluid proteins. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771(1–2):221–236. doi:10.1016/S1570-0232(02)00114-9
45. Pilzner C, Buhling F, Reinheckel T, et al. Allergic airway inflammation in mice deficient for the antigen-processing protease cathepsin E. Int Arch Allergy Immunol. 2012;159(4):367–383. doi:10.1159/000338288
46. Ladjemi MZ, Gras D, Dupasquier S, et al. Bronchial epithelial IgA secretion is impaired in asthma. Role of IL-4/IL-13. Am J Respir Crit Care Med. 2018;11:197.
47. Kang X, Li N, Wang M, et al. Adjuvant effects of ambient particulate matter monitored by proteomics of bronchoalveolar lavage fluid. Proteomics. 2010;10(3):520–531. doi:10.1002/pmic.200900573
48. Ousavi SA, Mohammadzadeh V, Loni E. MDetermination of alpha-1 antitrypsin level in patients with severe asthma. Tanaffos. 2013;12(4):19–22.
49. Feldman JM, Mayefsky L, Beckmann L, Lehrer PM, Serebrisky D, Shim C. Ethnic differences in asthma-panic disorder comorbidity. J Allergy Clin Immunol. 2010;125(3):760–762. doi:10.1016/j.jaci.2009.11.002
50. Haenen S, Clynen E, Vooght VD, et al. Proteome changes in auricular lymph nodes and serum after dermal sensitization to toluene diisocyanate in mice. Proteomics. 2012;12(23–24):3548–3558. doi:10.1002/pmic.201200264
51. Verrills NM, Irwin JA, He XY, et al. Identification of novel diagnostic biomarkers for asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183(12):1633–1643. doi:10.1164/rccm.201010-1623OC
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.