Back to Journals » Cancer Management and Research » Volume 12
DUS4L Silencing Suppresses Cell Proliferation and Promotes Apoptosis in Human Lung Adenocarcinoma Cell Line A549
Authors Li Z, Yin C, Li B, Yu QY, Mao WJ, Li J, Lin JP, Meng YQ, Feng HM, Jing T
Received 26 June 2020
Accepted for publication 17 September 2020
Published 9 October 2020 Volume 2020:12 Pages 9905—9913
DOI https://doi.org/10.2147/CMAR.S265671
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Zheng Li,* Ci Yin,* Bin Li, Qi-Yao Yu, Wen-Jie Mao, Jie Li, Jun-Ping Lin, Yu-Qi Meng, Hai-Ming Feng, Tao Jing
Department of Thoracic Surgery, Lanzhou University Second Hospital, Lanzhou University Second Clinical Medical College, Lanzhou 730030, China
*These authors contributed equally to this work
Correspondence: Bin Li
Department of Thoracic Surgery, Lanzhou University Second Hospital, Lanzhou University Second Clinical Medical College, Lanzhou 730030, China
Tel/Fax +86 931 8942073
Email [email protected]
Purpose: This study aims to investigate the potential role of DUS4L (dihydrouridine synthase 4 like) in lung adenocarcinoma (LUAD) and explore its associated pathways in human LUAD.
Methods: Firstly, we evaluated the relationships between clinicopathological characteristics and DUS4L expression via analysis of TCGA RNA sequencing data and other publicly available databases. Then, DUS4L was effectively silenced in LUAD cell line A549 using the lentiviral shRNA (short-hairpin RNA) transfection to assess its effects on cell proliferation, cycle and apoptosis in LUAD cells. RNA-seq technology was applied to shDUS4L and shCtrl-transfected cells to generate the corresponding gene expression profiles. Differentially expressed genes (DEGs) were identified using the DESeq2 program package. Also, DEGs were subjected to Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis to explore the associated molecular signaling pathways and relevant biological functions.
Results: Analysis of TCGA data revealed that DUS4L was highly upregulated in LUAD tissues which was related to clinical T and TNM stages of LUAD. The knockdown of DUS4L effectively inhibited cell proliferation and promoted apoptosis in A549 cells. Furthermore, the DEGs between the shDUS4L and shCtrl A549 cells were mainly enriched in biological processes associated with spliceosome, ribosome, RNA catabolic process, ncRNA (non-coding RNA) processing, and p53 signaling pathway.
Conclusion: Altogether, our results suggest that DUS4L is significantly associated with tumorigenesis and could be utilized as a novel biomarker and therapeutic target for LUAD.
Keywords: DUS4L, lung adenocarcinoma, proliferation, apoptosis, TCGA
Introduction
Cancer is one of the biggest public health challenges in the world.1 According to global data released by the International Agency for Research on Cancer (IARC), among all types of cancer, lung cancer is the leading cause of cancer-related death worldwide with nearly 2.1 million new cases of lung cancer and over 1.76 million mortalities in 2018, accounting for nearly one fifth of all cancer deaths.2 Moreover, the cases of lung adenocarcinoma (LUAD), one of the major histological subtypes of lung cancer, have been increasing over time and account for almost 50% of the total lung cancer patients.3,4 In the last few decades, the rapid development of medical and clinical treatment technologies such as chemotherapy, molecular targeted therapy, stereotactic radiotherapy, and immunotherapy saved thousands of lung cancer patients.5–7 However, due to the asymptomatic nature of the early disease, at the time of diagnosis, approximately 70% of the lung cancer patients are in advanced stages III–IV, having an average 5-year survival rate of less than 20%.8 Therefore, it is of utmost importance to advance the understanding of the regulatory mechanisms involved in the development and progression of LUAD. Also, more sensitive novel biomarkers need to be identified for the early diagnosis and therapeutic purposes.
Recently, it was shown that DUS4L (dihydrouridine synthase 4 like) plays an important role in the development of malignant tumor cells, including prostate and gastric cancers.9,10 However, its role in LUAD remains largely unknown. Therefore, in this study, using several publicly available databases, we investigated the role of DUS4L in LUAD. Also, its effects on cell proliferation and apoptosis were examined by carrying out the knockdown studies in the human LUAD cell line, A549. Furthermore, using the bioinformatics tools, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed to predict the associated molecular signaling pathways and relevant biological functions.
Materials and Methods
Bioinformatic Analysis
The RNA-seq mRNA expression profiles (Workflow Type: HTSeq-FPKM) of DUS4L in LUAD, including a total of 535 LUAD and 59 adjacent normal tissues, and the corresponding clinical data information were extracted from TCGA Genomic Data Commons (GDC) Portal (https://gdc.cancer.gov/). Data related to patients with missing clinical information and expression were excluded. UALCAN (http://ualcan.path.uab.edu/)11 and Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/index.html)12 databases were utilized to analyze DUS4L mRNA expression in non-paired samples of LUAD. Also, the Wilcoxon signed-rank test was performed to analyze DUS4L mRNA expression in 57 paired samples of LUAD. Lastly, the Mann–Whitney U-test was used to analyze the relationship between DUS4L mRNA expression and clinicopathological features of LUAD patients.
Cell Culture
The human lung adenocarcinoma cells A549 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum at 37°C and 5% CO2 using a standard humidified incubator. Also, the culture medium was changed every three days.
Lentiviral shRNA-Mediated Silencing of DUS4L
The short hairpin RNAs (shRNA) were synthesized by Genechem Chemtech (Shanghai, China). To design RNA interference sequences, the DUS4L gene (NM_181581) was used as a template. The sequences of shRNA targeting DUS4L and the negative control were 5ʹ-ACATCAGCAATCATAGATT-3ʹ and 5ʹ-TTCTCCGAACGTGTCACGT-3ʹ, correspondingly. After transfection, cells were harvested and the knockdown efficiency was determined using quantitative RT-PCR and Western blotting.
Western Blotting
Western blotting was used to analyze the cellular protein levels. Total cellular proteins were extracted using a lysis buffer and concentration was measured by the BCA Protein Assay kit (Beyotime Biotechnology, Shanghai, People’s Republic of China). Equal amounts of corresponding cell lysates were separated on a 10% sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE; Tanon Technology, Shanghai, China) and proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The PVDF membrane was blocked with Tris-buffered saline with Tween-20 (TBST) containing 5% non-fat milk for 1 hour at room temperature (RT) or overnight at 4°C. This was followed by incubation with the primary and then the secondary antibodies. The protein bands on membranes were visualized using the ECL Western Blotting Substrate kit (Pierce, Thermo Fisher Scientific) according to the manufacturer’s instructions. The primary anti-flag antibody was purchased from Sigma Biotechnology (Sigma-Aldrich Co., St Louis, MO, USA), anti-GAPDH, and secondary antibodies were obtained from Cruz Biotechnology Inc.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was reverse transcribed into cDNA using the M-MLV kit (Promega, Madison, Wisconsin, USA). LightCycler 480 II real-time PCR instrument (Roche Applied Science, Indianapolis, IN, USA) was used to perform the Quantitative real-time PCR (qRT-PCR). The sequences of the used primers were as follows: DUS4L: forward 5ʹ-GCCCATTGATTGTTCAGTTTGC-3ʹ, reverse 5ʹ-AACTCCTGTTGCTTCAGCCTTT-3ʹ; GAPDH: forward 5ʹ-TGACTTCAACAGCGACACCCA-3ʹ, reverse 5ʹ-CACCCTGTTGCTGTAGCCAAA-3ʹ. The corresponding expression levels were calculated using the formula 2−∆∆Ct.
Cell Growth Assay
2x103 cells/well were seeded into 96-well plates and incubated overnight at 37°C and 5% CO2. After plating, from the second day onwards, the number of cells was monitored once a day for the five consecutive days using a Celigo image cytometer (Nexcelom Bioscience, Lawrence, MA, USA). Cell growth curves were obtained using the corresponding fold change in cell proliferation.
Colony Formation Assay
The cells were seeded into 6-well plates and cultured for 15 days. After fixation with paraformaldehyde (4%), the colonies were stained with Crystal violet solution (1000 μL) for 15 minutes. Then, the cell colonies were photographed and counted under a microscope (Olympus).
Cell Cycle Assay
shDUS4L and shCtrl transfected cells were inoculated into 6-well plates and cultured for 48 hours. Then, cells were fixed with 75% ethanol for 1 h at 4°C, subsequently washed with D-Hanks solution (pH=7.2~7.4) (Genechem Co., LTD.) and collected. The fixed cells were stained with 2 mg/mL propidium iodide (PI) and subjected to cell cycle analysis using a Guava easyCyte HT flow cytometer (Millipore, USA).
Flow Cytometry Analysis
To examine cell apoptosis, the cells were cultured into 6-well plates. Upon reaching ~85% confluence, cells were collected and stained using the Annexin V-Allophycocyanin (APC) single-staining Apoptosis Detection kit (eBioscience, San Diego, CA, USA), as per the manufacturer’s instructions.
Enrichment Analysis
RNAi technology has been used widely for gene silencing. In this study, shRNA technology was employed to construct a DSU4L knockout in the A549 cell line. Subsequently, the gene expression data were generated using the RNA-seq technology. Program package DESeq213 was used to analyze the differential expression of RNAs in shCtrl and shDUS4L A549 cells. Genes were considered differentially expressed using the criterion of fold change >2 and P-value <0.05. GO and KEGG enrichment analysis were performed using the ClusterProfiler package (https://bioconductor.org/packages/release/clusterProfiler.html) in R software by adjusting the significance threshold of P-value <0.05.
Statistical Analysis
The statistical analysis was performed using the software SPSS version 18.0 for Windows. One-way ANOVA was applied for comparisons between multiple groups and the difference between the two groups was subjected to unpaired students’ t-tests. All data are expressed as mean ± SD and a P-value <0.05 was considered statistically significant.
Results
Bioinformatic Analysis
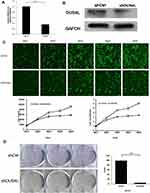
We analyzed the mRNA expression of DUS4L in LUAD using the TCGA data. We found that DUS4L expression was upregulated in lung cancer tissues compared to the non-paired (Figure 1A and B) or paired normal tissues (Figure 1C). Interestingly, TCGA data revealed that DUS4L upregulation was associated with clinical T and TNM stages of LUAD patients (Table 1). Overall, these results suggest that DUS4L is associated with the progression of LUAD.
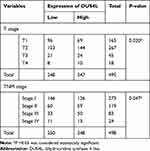 |
Table 1 Relationship Between DUS4L mRNA Expression and Clinical Pathological Parameters by TCGA |
DUS4L Knockdown Inhibits Proliferation of A549 Cells
To evaluate the effects of DUS4L on cell growth, lentivirus-mediated DUS4L knockdown was performed in A549 cells. This was verified using Western blotting and qRT-PCR. The results show that DUS4L was efficiently silenced in shDUS4L A549 cells (Figure 2A and B). Furthermore, cell growth assay showed that compared to the shCtrl group, DUS4L knockdown inhibited the proliferation of A549 cells (Figure 2C). Also, the colony numbers were reduced in shDUS4L A549 cells compared to the shCtrl cells (Figure 2D). Collectively, these results show that DUS4L promotes the proliferation and colony formation of A549 cells.
Effect of DUS4L on Cell Cycle and Cell Apoptosis
As shown in Figure 3A, compared with the shCtrl group, the shDUS4L group showed a marked increase in the number of cells in the G2/M-phase phase. Also, the number of cells in the G1-phase decreased. This suggests that the DUS4L is associated with the periodic distribution of LUAD cells. Moreover, the number of apoptotic A549 cells in the shDUS4L group increased significantly compared to the shCtrl group (Figure 3B).
GO and KEGG Enrichment Analysis
To explore the role of DUS4L in LUAD, we identified the differential expressed genes (DEGs) in the shCtrl and shDUS4L A549 cells using the RNA-seq technology. We found that knockdown of DUS4L up-regulated 2206 genes while 2808 genes were down-regulated (Figure 4). GO (Table 2) and KEGG (Table 3) enrichment analysis revealed that the DEGs were significantly enriched in pathways related to spliceosome, ribosome, RNA catabolic process, ncRNA processing, p53 signaling, and so on.
 |
Table 2 GO Enrichment Analysis for Differentially Expressed Genes |
 |
Table 3 KEGG Enrichment Analysis for Differentially Expressed Genes |
 |
Figure 4 Heat map for differentially expressed genes from RNA-seq profile (D). |
Discussion
Using the Gene Cards Database and UCSC Genome Browser, we found that the human DUS4L gene, also named as DUS4 or PP35, is located in chromosome 7q22.3, spanning eight regions. DUS4L itself was rarely associated with human cancers, but Kim HP9 and NacuS10 discovered that chimeric RNA DUS4L-BCAP29, composed of DUS4L exon fragments and BCAP29, was a tumor-promoting factor in prostate and gastric cancers. Later, Tang et al14–16 showed that DUS4L-BCAP29 fusion transcript exists in multiple non-neoplastic tissues and cells, including prostate and gastric cancers, and can effectively promote hUC-MSCs (human umbilical cord mesenchymal stem cells) proliferation and differentiation into neuron-like cells. Interestingly, several other studies17–19 reported that DUS4L was also associated with the onset of osteoarthritis.
To date, no report has associated DUS4L to LUAD. For the first time, our study examined the expression of DUS4L in human LUAD using bioinformatic methods. We found that DUS4L mRNA expression, by comparing paired tumor and adjacent samples and non-paired samples, was significantly elevated in the tumor tissues. Additionally, the upregulation of DUS4L was associated with tumor size and TNM stages in LUAD. Therefore, we wanted to test whether altered expression of DUS4L would affect the proliferation and apoptosis of LUAD cells. After efficiently establishing the null expression of DUS4L via lentiviral-mediated knockdown, we found that DUS4L knockdown could extensively inhibit the proliferation of LUAD A549 cells and induce cell cycle arrest while promoting cell apoptosis. However, the potential molecular mechanisms forcing such changes need to be elucidated. Transcriptomics analysis revealed that DUS4L knockout significantly altered the gene expression profile of A549 cells. A total of 5011 DEGs were screened, of which 2206 genes were up-regulated and 2805 genes were down-regulated. These DEGs were enriched in vital biological processes such as spliceosome, ribosome, RNA catabolic process, ncRNA processing, and p53 signaling pathway. Notably, several studies have demonstrated that dysfunction of the spliceosome,20,21 ribosomal proteins,22,23 small non-coding RNAs, including transfer RNAs (tRNAs),24,25 and microRNAs (miRNAs)26 play an important role in tumorigenesis.
However, the present study had some limitations. This study established the role of DUS4L in LUAD by examining its mRNA expression levels. It would be interesting to examine the changes also at the protein level. Moreover, the role of DUS4L needs to be verified using the clinical samples. Nonetheless, this study set the platform for future investigations which could help in understanding the specific mechanisms behind DUS4L mediated regulations.
Conclusion
In conclusion, for the first time, our study reports that DUS4L is upregulated in LUAD which increases LUAD cell growth in vitro. Moreover, these results suggest that DUS4L should be considered an oncogene in LUAD which can be exploited as a novel therapeutic target.
Acknowledgments
This study was supported by Cuiying Graduate Supervisor Applicant Training Program of Lanzhou University Second Hospital (Grant ID: 201801) and Technological Development Contract of Lanzhou University (Grant ID: (19)0402).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):
3. Yatabe Y, Borczuk AC, Powell CA. Do all lung adenocarcinomas follow a stepwise progression? Lung Cancer. 2011;74(1):7–11. doi:10.1016/j.lungcan.2011.05.021
4. Ladanyi M, Pao W. Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond. Mod Pathol. 2008;21(Suppl 2):S16–S22. doi:10.1038/modpathol.3801018
5. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi:10.1038/nature25183
6. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8
7. Mathew M, Enzler T, Shu CA, Rizvi NA. Combining chemotherapy with PD-1 blockade in NSCLC. Pharmacol Ther. 2018;186:
8. Moumtzi D, Lampaki S, Zarogoulidis P, et al. Prognostic factors for long term survival in patients with advanced non-small cell lung. Ann Transl Med. 2016;4(9):161. doi:10.21037/atm.2016.05.13
9. Kim HP, Cho GA, Han SW, et al. Novel fusion transcripts in human gastric cancer revealed by transcriptome analysis. Oncogene. 2014;33(47):
10. Nacu S, Yuan W, Kan Z, et al. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med Genomics. 2011;4(1):11. doi:10.1186/1755-8794-4-11
11. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi:10.1093/nar/gkz430
12. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
13. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
14. Tang Y, Qin F, Liu A, Li H. Recurrent fusion RNA DUS4L-BCAP29 in non-cancer human tissues and cells. Oncotarget. 2017;8(19):
15. Tang Y, Ma S, Wang X, et al. Identification of chimeric RNAs in human infant brains and their implications in neural differentiation. Int J Biochem Cell Biol. 2019;111:
16. Tang Y, Guan F, Li H. Case study: the recurrent fusion RNA DUS4L-BCAP29 in noncancer human tissues and cells. Methods Mol Biol. 2020;2079:
17. Raine EV, Wreglesworth N, Dodd AW, Reynard LN, Loughlin J. Gene expression analysis reveals HBP1 as a key target for the osteoarthritis susceptibility locus that maps to chromosome 7q22. Ann Rheum Dis. 2012;71(12):
18. Evangelou E, Valdes AM, Kerkhof HJ, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis. 2011;70(2):
19. Hämäläinen S, Solovieva S, Vehmas T, Luoma K, Leino-Arjas P, Hirvonen A. Genetic influences on hand osteoarthritis in finnish women–a replication study of candidate genes. PLoS One. 2014;9(5):e97417. doi:10.1371/journal.pone.0097417
20. Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212(1):
21. Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64(21):
22. Gou Y, Shi Y, Zhang Y, et al. Ribosomal protein L6 promotes growth and cell cycle progression through upregulating cyclin E in gastric cancer cells. Biochem Biophys Res Commun. 2010;393(4):
23. Cheng DD, Zhu B, Li SJ, Yuan T, Yang QC, Fan CY. Down-regulation of RPS9 inhibits osteosarcoma cell growth through inactivation of MAPK signaling pathway. J Cancer. 2017;8(14):
24. Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37(21):
25. Zhang Z, Ye Y, Gong J, et al. Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers. Commun Biol. 2018;1:234. doi:10.1038/s42003-018-0239-8
26. Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.