Back to Journals » Journal of Hepatocellular Carcinoma » Volume 8
DTYMK Expression Predicts Prognosis and Chemotherapeutic Response and Correlates with Immune Infiltration in Hepatocellular Carcinoma
Authors Guo Y , Luo W, Huang S , Zhao W, Chen H, Ma Y, Ye M, Nie Y, Zhang Y, Huang C, Zhou Q, He X, Chen M
Received 24 March 2021
Accepted for publication 6 July 2021
Published 3 August 2021 Volume 2021:8 Pages 871—885
DOI https://doi.org/10.2147/JHC.S312604
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Ahmed Kaseb
Yiwen Guo,1– 3,* Weixin Luo,1– 3,* Shanzhou Huang,4,* Wenting Zhao,5 Huadi Chen,1– 3 Yihao Ma,1– 3 Maodong Ye,1– 3 Yu Nie,1– 3 Yixi Zhang,6 Changjun Huang,1– 3 Qi Zhou,7,8 Xiaoshun He,1– 3 Maogen Chen1– 3
1Organ Transplant Center, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, 510080, People’s Republic of China; 2Guangdong Provincial Key Laboratory of Organ Donation and Transplant Immunology, Guangzhou, 510080, People’s Republic of China; 3Guangdong Provincial International Cooperation Base of Science and Technology (Organ Transplantation), Guangzhou, 510080, People’s Republic of China; 4Department of General Surgery, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, 510080, People’s Republic of China; 5School of Traditional Chinese Medicine, Southern Medical University, Guangdong, Guangzhou, 510515, People’s Republic of China; 6Liver Transplantation Center, Beijing Friendship Hospital, Capital Medical University, Beijing, 100005, People’s Republic of China; 7Department of General Surgery, Hui Ya Hospital of The First Affiliated Hospital, Sun Yat-sen University, Huizhou, Guangdong, 516081, People’s Republic of China; 8Department of Liver Surgery, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, 510080, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Maogen Chen; Xiaoshun He
Organ Transplant Center, The First Affiliated Hospital, Sun Yat-sen University, No. 58 Zhongshan Er Road, Guangzhou, 510080, People’s Republic of China
Tel +86-18122731962
; +86-20-87306082
Fax +86-20-87306082
Email [email protected]; [email protected]
Introduction: Hepatocellular carcinoma (HCC) is the most common malignant tumor of the liver. Identifying specific molecular markers that can predict HCC prognosis is extremely important. The protein deoxythymidylate kinase (DTYMK) has been reported to contribute to unfavorable prognosis in non-small cell lung cancer patients, but its role in the prediction of HCC patient prognosis has not been clarified.
Methods: Samples from the TCGA and GEO databases were consecutively enrolled for gene expression analysis, clinicopathology analysis, immune microenvironment analysis and chemotherapeutic response prediction. The results were validated using 86 samples from the First Affiliated Hospital of Sun Yat-sen University. Cox regression analysis was used to analyze the effect of DTYMK on progression-free survival (PFS) and overall survival (OS). Functional enrichment analysis was used to describe the marker pathways that were significantly related to DTYMK. TIMER (Tumor Immune Estimation Resource), TISIDB (Tumor and Immune System Interaction DataBase) and CIBERSORT (Cell type Identification By Estimating Relative Subsets Of RNA Transcripts) were used to explore the immune microenvironment.
Results: We found that DTYMK expression upregulation is associated with poor prognosis in HCC patients and tightly related to the pathways regulating the cell cycle and acid metabolism. Our findings revealed that hepatocellular carcinoma cell lines with high DTYMK expression were more sensitive to sorafenib and many other chemotherapeutic drugs. We also found an inhibiting effect of DTYMK on the immune microenvironment in the process of tumorigenesis.
Discussion: We found that DTYMK has potential as a new prognostic and chemotherapeutic response biomarker for HCC patients and correlates with the immune microenvironment in HCC. However, there are some deficiencies in our study. First, this is a retrospective study that may lead to selection bias. Second, the protein expression of DTYMK was investigated via immunohistochemical analysis. Finally, we did not explore the exact underlying molecular mechanisms of DTYMK in tumorigenesis in this study, which is needed to be clarified in future research.
Keywords: hepatocellular carcinoma, DTYMK, cell cycle, immune infiltration, TCGA
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide, leading to approximately 500,000 deaths worldwide each year.1,2 HCC is the leading histological type of primary liver cancer (PLC) and comprises 75%-85% of PLCs. In the past, hepatitis virus infection, aflatoxin intake and excessive drinking were the main etiological factors of HCC, but recent studies have revealed that obesity-related nonalcoholic fatty liver disease and nonalcoholic steatohepatitis are also major causes of HCC.3,4 At present, there are multiple therapeutic methods available for HCC, including radical resection or liver transplantation. However, because liver cancer is often in the advanced stages by the time many patients seek medical advice, the prognosis of HCC patients is still unfavorable.5 Hence, efficient biomarkers are vital for identifying early HCC.
Deoxythymidylate kinase (DTYMK), a nuclear deoxythymidylate kinase, catalyzes the process of deoxy-TMP phosphorylation. DTYMK is widely expressed in all tissues and catalyzes the last reaction of the deoxyribonucleoside triphosphate (dTTP) production pathways.6 Previous studies have shown that DTYMK is probably a predictive factor of poor prognosis in non-small cell lung cancer (NSCLC) and may be a potential therapeutic target in NSCLC.7–9 Yeh et al mentioned that DTYMK might have relevance to the poor prognosis of HCC patients,10 but its importance has not yet been evaluated.
This is the first elaborate analysis of the relationship between DTYMK and HCC development. In this study, we used samples data The Cancer Genome Atlas (TCGA) and our cohort to investigate the role of DTYMK expression on the prognosis of HCC patients. We assessed the transcriptional changes in DTYMK in HCC and the impact of these changes on tumor immune cell infiltration and the chemotherapeutic response. These results may provide a novel diagnostic and therapeutic strategy for HCC patients.
Method
Patients and Gene Expression Profile
HCC patients with RNA sequencing and clinicopathological data available from the TCGA database (https://gdc.cancer.gov/) were enrolled for the analyses. Sample data from the GEO database (GSE14520, GSE25097, GSE63898) (http://www.ncbi.nlm.nih.gov/geo) were also analyzed. According to the median mRNA expression level of DTYMK, the patients from the TCGA were divided into high and low expression groups. We also recruited 86 HCC patients without preoperative radiotherapy or chemotherapy from the First Affiliated Hospital of Sun Yat-Sen University as a validation cohort. In accordance with the immunohistochemistry score of DTYMK, the patient cohort was also divided into two groups.
Gene Set Enrichment Analysis
Gene set enrichment analysis(GSEA) was used to predict possible GO and KEGG pathways related to DTYMK expression using RNA-seq data from the TCGA database. We performed 1000 gene set permutations times for each analysis. Enrichment with a false discovery rate < 0.05 and a nominal p-value < 0.05 was considered statistically significant.
Immune Infiltration Analysis
The relative fraction of 22 types of tumor-infiltrating immune cells (TIICs) in HCC patients from the TCGA database was calculated by CIBERSORT.11 We compared the relative abundance of TIICs between the high and low DTYMK expression groups. The Tumor Immune Estimation Resource (TIMER)12 was used to conduct integrated correlation analysis between tumor-infiltrating immune cell signatures and DTYMK expression levels. Based on an integrated repository portal for tumor-immune system interactions (TISIDB),13 we explored the relation between DTYMK expression levels and immune-related molecules.
Chemotherapeutic Response Prediction
Chemotherapeutic response was predicted for the TCGA samples based on the Genomics of Drug Sensitivity in Cancer database (GDSC, https://www.cancerrxgene.org/). Eighteen selected chemotherapeutic drugs commonly used for HCC, including sorafenib and others, were evaluated. The R package “pRRophetic”14 was applied, and the concentration needed to inhibit the growth of half of the cells (IC50) was estimated by a ridge regression model, which reflected the drug sensitivity.
Immunohistochemistry Staining
From our validation cohort, 86 formalin-fixed paraffin-embedded slides were deparaffinated, hydrated, blocked and mixed with the primary anti-DTYMK polyclonal antibody (Abcam, ab241493) and incubated overnight at 4°C. Finally, HCC tissue staining was conducted and assessed as previously described.15
Statistical Analysis
All statistical analyses were performed in R (Version 3.6.1). The Wilcoxon rank sum test was used to compare the difference between two groups with quantitative data. The overall and disease-free survival curves were assessed by Kaplan-Meier analysis and compared by a two-sided Log rank test. The relationship between clinical characteristics and DTYMK expression was analyzed by Fisher’s exact test. Univariate and multivariate analyses were performed by Cox regression models to find independent variables related to prognosis. A P value less than 0.05 was defined as statistically significant.
Ethics Approval
The study was reviewed and approved by the Institutional Review Board of The First Affiliated Hospital of Sun Yat-Sen University and conformed to the tenets of the Declaration of Helsinki. Written informed consent was obtained from the patients.
Result
DTYMK Expression Upregulation is Associated with Poor Prognosis in HCC Patients
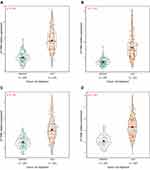
To determine the expression of DTYMK in HCC tissues, we first analyzed 631 adjacent samples and 721 HCC samples in three GEO datasets (GSE14520, GSE25097, and GSE63898). The results demonstrated that, compared with tumor adjacent tissues, DTYMK expression in HCC tissues was significantly upregulated (Figure 1A–C, p all <0.001), which was in line with the expression data from the TCGA database cohort (Figure 1D, p <0.001).
We analyzed the relationship between the expression of DTYMK and clinicopathological features in HCC patients from the TCGA database (Table 1). The results indicated that the DTYMK expression level was significantly associated with primary tumor size, disease stage and pathological grade. As shown in Table 2, we further explored connections between DTYMK expression and overall survival (OS) or disease-free survival (DFS) and other clinicopathological characteristics in HCC patients. Univariate analysis showed that there were four factors associated with HCC OS (disease stage, tumor size, metastasis and expression level group) and three factors associated with HCC DFS (disease stage, tumor size and expression level group) (Table 2A). Our multivariate analysis demonstrated that the DTYMK expression level (p<0.05) was an independent prognostic factor. Figure 2A shows the distribution of DTYMK expression and the survival status of HCC patients. The AUC of the DTYMK expression level for forecasting overall survival was 0.633 (Figure 2B). We divided HCC patients from the TCGA database into two groups based on DTYMK expression level, and survival analysis was further performed. As shown in Figure 2C and D, the expression level of DTYMK was tightly related to patient survival and disease recurrence.
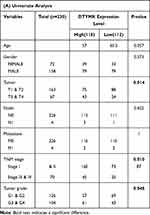 |
Table 1 Correlation Between DTYMK Expression Level and Clinicopathological Features of HCC in TCGA Database |
 |
Table 2 Univariate or Multivariate Analysis of OS/DFS and Clinicopathological Parameters in HCC Patients |
GSEA Investigation of DTYMK
To understand the possible biological functions of DTYMK, we performed GO term enrichment and KEGG pathway analysis by the GSEA method. The results revealed that the DTYMK expression level was significantly different with an FDR<0.05 and a p-value<0.05 in the pathway enrichment analysis. As shown in Table 3A, GO analysis showed that protein activation cascades, monooxygenase activity, androgen metabolism, organic acid catabolism and monocarboxylic acid catabolism were negatively correlated with DTYMK; DNA biosynthesis, checkpoint signal transduction involved in the cell cycle, condensed chromosome centromeric region and negative cell cycle regulation were positively correlated with DTYMK. As shown in Figure 3A and B, cell cycle progression and acid catabolism were the key biological processes and were closely associated with DTYMK. Meanwhile, KEGG pathway analysis uncovered five terms negatively correlated with high levels of DTYMK, fatty acid metabolism, complement and coagulation cascades, drug metabolism cytochrome p450, valine leucine and isoleucine degradation and tryptophan metabolism. Five positively associated pathways were also confirmed, including base excision repair, pyrimidine metabolism, homologous recombination, DNA replication and the cell cycle (Table 3B–D). In summary, DTYMK expression was tightly related to the pathways regulating the cell cycle and acid metabolism, which are crucial in HCC.
 |
Table 3 Signaling Pathways Most Strongly Associated with DTYMK Expression Level Based on Their Normalized Enrichment Score (NES) and p-value |
Profiles of 22 Tumor Infiltrating Immune Cells (TIICs) in HCC
First, to investigate the potential interaction among different immune cell types infiltrating HCC, we computed the correlations between 22 immune cell types and the CIBERSORT p-values. Some immune cell types had a potential relationship in the TCGA cohort (Figure 4A). The most relevant cells were resting mast cells and activated mast cells with a negative R value of −0.65. Naïve B cells and memory B cells also had a negative R value of −0.58. Interestingly, however, there was a moderate correlation between regulatory T cells and resting NK cells with an R value of −0.47. These results suggested that mast cells and humoral immunity are critical in the pathogenesis of HCC.
We next analyzed the distribution of the 22 TIICs in different DTYMK expression level groups. As shown in Figure 4B, follicular helper T cells (Tfhs), regulatory T cells (Tregs) and M0 macrophages were significantly different between the high- and low-expression groups, which implied the importance of Tfhs, Tregs and M0 macrophages. Moreover, the largest difference was found in M2 macrophages in both groups, suggesting a critical role for these cells in tumor progression.
Then, we evaluated the relationship between DTYMK expression and immune infiltration levels using TIMER. As illustrated in Figure 4C, the expression level of DTYMK was positively correlated with tumor purity (r=0.139, p=9.50e-03). Moreover, there was a positive relationship between DTYMK expression and the infiltration levels of CD4+ T cells (r=0.314, p=2.56e-09), B cells (r=0.364, p=2.83e-12), macrophages (r=0.303, p=8.99e-09), myeloid dendritic cells (r=0.46, p=1.64e-19) and neutrophils (r=0.3009, p=4.51e-09) in HCC. It is worth noting that the correlation between DTYMK and CD8+ T cells was not significant.
Finally, we verified the correlation between DTYMK and immune-related molecules in 373 sample from patients with liver hepatocellular carcinoma (LIHC) by using the TISIDB database. As illustrated in Figure 5, eight immunostimulatory molecules and five immunosuppressive molecules were correlated with DTYMK expression. Specifically, DTYMK expression had positive or negative correlations with stimulatory molecules, such as CD28 (r=−0.137, p=7.99e-03), CD40LG (r=−0.137, p=8.07e-03), CXCL12 (r=−0.301, p=3.49e-09), ICOS (r=0.132, p=1.08e-02), ICOSLG (r=−0.126, p=1.48e-02), IL-6 (r=−0.229, p=8.6e-06), TNFSF9 (r=−0.162, p=1.71e-03) and TNFSF19 (r=−0.283, p=3.06e-08). Similarly, five inhibitory molecules were correlated with DTYMK, CD160 (r=0.13, p=1.23e-02), CTLA4 (r=0.223, p=1.51e-05), LAG3 (r=0.257, p=5.4e-07), PDCD1 (r=0.173, p=8.08e-04) and CD274 (r=−0.264, p=2.64e-07). In addition to these molecules, some molecules tested for in the LIHC cohort had no significant correlation with DTYMK (Figure S1).
 |
Figure 5 (A) Correlation analysis of DTYMK and immunostimulatory molecules. (B) Correlation analysis of DTYMK and immunosuppressive molecules. |
More Sensitivity to Chemotherapeutic Drugs in the High DTYMK Expression Group
Chemotherapy is a critical therapy for patients with later-stage HCC, and we evaluated the response of high and low DTYMK expression human hepatocellular carcinoma cell lines to 18 chemotherapeutic drugs (Table S1), such as sorafenib and cisplatin, by applying the “pRRophetic” R package. Interestingly, the high DTYMK expression group showed more sensitivity to 12 of the 18 chemo drugs, with a lower IC50 (Figure 6 and Figure S2). This finding gives physicians a hint about offering HCC patients the individualized therapy.
Data Validation Based on Our Cohort
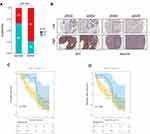
Based on our patient cohort of 86 patients with HCC, immunohistochemistry staining results showed that 63 had high expression of DTYMK, while 23 had low expression (Figure 7A and B). DTYMK protein expression was obviously upregulated in HCC compared with normal tissue. We checked the relevance between the expression of DTYMK and clinicopathological features in HCC patients from our cohort (Table 4). We found that DTYMK expression correlated with the pathological differentiation grade. Univariate and multivariate Cox regression analyses showed that DTYMK expression was an independent risk factor for prognosis in HCC patients (Table 5). In addition, a significant validated correlation existed between DTYMK expression and poor OS and DFS rates by Kaplan-Meier survival analysis (Figure 7C and D).
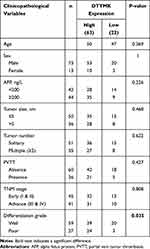 |
Table 4 Correlation Between DTYMK Expression and Clinicopathological Features of HCC in Our Validation Cohort |
 |
Table 5 Univariate or Multivariate Analysis of OS/DFS and Clinicopathological Parameters in HCC Patients from Our Validation Cohort |
Discussion
Currently, very little is known about the role of DTYMK in cancer. Only a few reports have shown that high expression of DTYMK is closely correlated with poor prognosis in patients with LKB1 mutant NSCLC.7–9 In these studies, Liu et al hypothesized that DTYMK could serve as a therapeutic target for LKB1 mutation-associated NSCLC. Hence, we designed the present study with the aim of investigating the role of DTYMK in HCC and its potential correlation with tumorigenesis and the prognosis of HCC patients.
First, we found that the DTYMK expression level in HCC tissues was significantly higher than that in adjacent normal tissues by analyzing the data from the GEO and TCGA databases. In the current study, we demonstrated that DTYMK might serve as a prognostic biomarker in HCC patients. The prognostic value of DTYMK was validated using data from the TCGA, and survival probability was predicted using Kaplan-Meier analysis. Poorly differentiated liver cancer was more likely to exhibit high expression of DTYMK. To validate this conclusion, 86 pairs of HCC and adjacent normal tissue samples were selected from our center. Immunochemical results showed that the difference in DTYMK expression between the HCC tissues and adjacent normal tissues was significant, and DTYMK levels in tumor tissues were higher than those in adjacent normal tissues. The above results implied the critical role of DTYMK in HCC progression and development.
We carried out further study to investigate the function of DTYMK in HCC by performing GSEA using the TCGA data. GO term and KEGG pathway analyses suggested that upregulated DTYMK expression was closely related to the cell cycle and acid metabolism in cancer. The results showed that DNA biosynthesis, condensed chromosome centromeric region, signal transduction involved in the cell cycle, checkpoint negative regulation of the cell cycle in GO and base excision repair, pyrimidine metabolism, homologous recombination, DNA replication, and cell cycle in KEGG were differentially enriched in tissues with high DTYMK expression. These results all indicated that DTYMK has the potential to be a prognostic marker and therapeutic target for HCC patients.
Furthermore, we revealed the connection between DTYMK expression and immune infiltration levels in HCC tissues using the TIMER database. The heatmap of 22 immune infiltrating cells in HCC samples suggested that Tregs were correlated with resting NK cells. Our CIBERSORT analysis showed a positive correlation between DTYMK expression and immune cell infiltration, particularly Tfhs, Tregs and M0 macrophages. HCC patients with high DTYMK expression had a higher infiltration level of Tregs, which caused impaired functional activity of NK cells. Robinson et al reported that NK cells could release cytotoxic granules to kill tumor cells.16 In addition, NK cells inhibit angiogenesis and tumor cell proliferation by secreting inflammatory cytokines.17 These findings revealed that Tfh cells, Tregs and M0 macrophages had higher infiltration levels in the high DTYMK expression group, which indicated a potential regulatory pathway of DTYMK on the function of T cells and macrophages in HCC. In addition, we found close links between DTYMK and CD4+ T cells, B cells, and myeloid dendritic cells, implying a possible effect of DTYMK on those immune cells in the process of tumorigenesis.
In addition to immune cell infiltration, we investigated the correlation between immune-related molecules and DTYMK. As seen in our results, DTYMK had a negative association with most immunostimulatory molecules and a positive association with most immunosuppressive molecules, suggesting that DTYMK might promote tumor progression by inhibiting antitumor immunity. The results revealed that the immunostimulatory molecules CXCL12, IL6, and TNFSF13 and the immunosuppressive molecules CTLA4, LAG3, and CD274 were the most strongly associated with DTYMK expression. As CTLA4 and LAG3 are immunosuppressive molecules, their high expression has been studied in relation to HCC patient prognosis,18,19 and previous findings are consistent with our results. DTYMK may influence the prognosis of HCC patients by modulating the expression levels of CTLA4 and LAG3. However, the expression levels of CXCL12, IL6, TNFSF13 and CD274 found here are inconsistent the levels reported in previous studies.20–24 In tissues with high expression of DTYMK, we found that the expression of CXCL12, IL6, TNFSF13 and CD274 was downregulated. Previous reports showed that these molecules were related to poor prognosis.22,25–27 Therefore, DTYMK may affect the prognosis of HCC through other molecular pathways. However, the mechanism by which DTYMK promotes tumor development by inhibiting antitumor immunity still needs more investigation.
Sorafenib obviously improves patient prognosis and was approved for treating advanced HCC in 2007;28 however, not all patients had a positive response to the mechanism of the drug. Our findings revealed that hepatocellular carcinoma cell lines with high DTYMK expression were more sensitive to sorafenib and many other chemotherapeutic drugs. It should be noted that these findings do not contradict with our finding that DTYMK expression upregulation is associated with poor prognosis in HCC patients. Firstly, the data of these HCC patients were obtained from the TCGA database. However, there are no available data concerning whether these HCC patients had received one or more drugs of those 18 chemotherapeutic drugs analyzed or not. Secondly, the data of those hepatocellular carcinoma cell lines were obtained from the GDSC database. So the origins of the data in these two seemingly opposing facts are different and these two facts should not be simply combined into one conclusion. In 2014, Yu et al developed circulating tumor cells cultures from patients for individualized testing of drug susceptibility.29 In the future, DTYMK expression may serve as a marker to predict chemotherapeutic response in the clinical setting by evaluating the response of high and low DTYMK expression hepatocellular carcinoma cell lines from patients to chemotherapeutic drugs.
However, there are some deficiencies in our study. First, this is a retrospective study that may lead to selection bias. Second, the protein expression of DTYMK was investigated via immunohistochemical analysis. Finally, we did not explore the exact underlying molecular mechanisms of DTYMK in tumorigenesis in this study, which is needed to be clarified in future research.
Conclusions
This is the first study to evaluate the role of DTYMK in HCC. DTYMK expression upregulation is associated with poor prognosis in HCC patients and tightly related to the pathways regulating the cell cycle and acid metabolism. Our findings revealed that hepatocellular carcinoma cell lines with high DTYMK expression were more sensitive to sorafenib and many other chemotherapeutic drugs. We also found an inhibiting effect of DTYMK on the immune microenvironment in the process of tumorigenesis.
Data Sharing Statement
The datasets used and/or analyzed in this study are available from the corresponding authors on reasonable request.
Acknowledgments
Supported by grants as follows: National Natural Science Foundation of China (81401324 and 81770410), the Guangdong Provincial Key Laboratory Construction Projection on Organ Donation and Transplant Immunology (2013A061401007, 2017B030314018 and 2020B1212060026), Special Funding from Guangdong Provincial People's Hospital(2020bq09, 8200100290,KY012021164), Collaborative Funding of Basic and Applied Basis Research Funding of Guangdong Province(2021A1515110536), Basic and applied basic research funding Guangdong Province(2021A1515011473, 2021A1515012441), the Science and Technology Program of Guangzhou(No. 202102020030), the Science and Technology Program of Huizhou(2019C0602009), the Science and Technology Program of Huizhou Daya Bay(2020030401), Funding for the Construction of Key Specialty in Huizhou(Qi Zhou).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gao Q, Zhu H, Dong L, et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. cell. 2019;179(2):561–577. doi:10.1016/j.cell.2019.08.052
2. Zhang Q, He Y, Luo N, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. cell. 2019;179(4):829–845. doi:10.1016/j.cell.2019.10.003
3. Petrick JL, Braunlin M, Laversanne M, et al. International trends in liver cancer incidence, overall and by histologic subtype,1978–2007. Int J Cancer. 2016;139(1534–1545). doi:10.1002/ijc.30211.
4. Villanueva A, Longo DL. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi:10.1056/NEJMra1713263
5. Allaire M, Nault J-C. Advances in management of hepatocellular carcinoma. Curr Opin Oncol. 2017;29(4):288–295. doi:10.1097/CCO.0000000000000378
6. Caspi R, Altman T, Dreher K, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi:10.1093/nar/gkr1014
7. Wang W, Guo ZH, Lu XP, et al. Elevated expression of DTYMK is associated with poor prognosis in patients with Non-small cell lung cancer. Int J Clin Exp Med. 2016;9(11):22027–22033.
8. Liu Y, Marks K, Cowley GS, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov. 2013;3(8):870–879. doi:10.1158/2159-8290.CD-13-0015
9. Liu Y, Cowley GS, Cohoon TJ, Root DE, Kwok-kin W. Identification of DTYMK and CHEK1 as therapeutic targets in LKB1 mutant non-small cell lung cancer. Cancer Res. 2012;72.
10. Yeh HW, Lee SS, Chang CY, Hu CM, Jou YS. Pyrimidine metabolic rate limiting enzymes in poorly-differentiated hepatocellular carcinoma are signature genes of cancer stemness and associated with poor prognosis. Oncotarget. 2017;8(44):77734–77751. doi:10.18632/oncotarget.20774
11. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
12. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307
13. Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics (Oxford, England). 2019;35(20):4200–4202.
14. Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9):e107468. doi:10.1371/journal.pone.0107468
15. Huang S, Li J, Tam NL, et al. Uridine-cytidine kinase 2 upregulation predicts poor prognosis of hepatocellular carcinoma and is associated with cancer aggressiveness. Mol Carcinog. 2019;58(4):603–615. doi:10.1002/mc.22954
16. Robinson MW, Harmon C, O’Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell Mol Immunol. 2016;13(3):267–276. doi:10.1038/cmi.2016.3
17. Muller-Hermelink N, Braumüller H, Pichler B, et al. TNFR1 signaling and IFN-gamma signaling determine whether T cells induce tumor dormancy or promote multistage carcinogenesis. Cancer Cell. 2008;13:507–518. doi:10.1016/j.ccr.2008.04.001
18. Rowshanravan B, Halliday N, Sansom DM. CTLA-4: a moving target in immunotherapy. Blood. 2018;131(1):58–67. doi:10.1182/blood-2017-06-741033
19. Yarchoan M, Xing D, Luan L, et al. Characterization of the immune microenvironment in Hepatocellular Carcinoma. Clin Cancer Res. 2017;23(23):7333–7339. doi:10.1158/1078-0432.CCR-17-0950
20. Xu Y, Fang F, Jiao H, et al. Activated hepatic stellate cells regulate MDSC migration through the SDF-1/CXCR4 axis in an orthotopic mouse model of hepatocellular carcinoma. Cancer Immunol Immunother. 2019;68(12):1959–1969. doi:10.1007/s00262-019-02414-9
21. Iwahasi S, Rui F, Morine Y, et al. Hepatic stellate cells contribute to the tumor malignancy of hepatocellular carcinoma through the IL-6 pathway. Anticancer Res. 2020;40(2):743–749. doi:10.21873/anticanres.14005
22. Chen L, Guan H, Gu C, Cao Y, Shao J, Wang F. miR-383 inhibits hepatocellular carcinoma cell proliferation via targeting APRIL. Tumour Biol. 2016;37(2):2497–2507. doi:10.1007/s13277-015-4071-1
23. Langhans B, Nischalke HD, Krämer B, et al. Role of regulatory T cells and checkpoint inhibition in hepatocellular carcinoma. Cancer Immunol Immunother. 2019;68(12):2055–2066. doi:10.1007/s00262-019-02427-4
24. Ma L, Feng F-L, Dong L-Q, et al. Clinical significance of PD-1/PD-Ls gene amplification and overexpression in patients with hepatocellular carcinoma. Theranostics. 2018;8(20):5690–5702. doi:10.7150/thno.28742
25. Li JH, Ma WJ, Wang GG, et al. Prognostic value of programmed cell death Ligand 1 (PD-L1) in patients with Hepatocellular Carcinoma: a meta-analysis. Front Immunol. 2018;9:2077. doi:10.3389/fimmu.2018.02077
26. Semaan A, Dietrich D, Bergheim D, et al. CXCL12 expression and PD-L1 expression serve as prognostic biomarkers in HCC and are induced by hypoxia. Virchows Arch. 2017;470(2):185–196. doi:10.1007/s00428-016-2051-5
27. Lai SC, Su YT, Chi CC, et al. DNMT3b/OCT4 expression confers sorafenib resistance and poor prognosis of hepatocellular carcinoma through IL-6/STAT3 regulation. J Exp Clin Cancer Res. 2019;38(1):474. doi:10.1186/s13046-019-1442-2
28. Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–390. doi:10.1056/NEJMoa0708857
29. Yu MBA, Aceto N, Bersani F, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Cancer Ther. 2014;345(6193):216–220.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.