Back to Journals » Infection and Drug Resistance » Volume 15
Drug Resistance and Homology of Pathogens of Infective Diarrhea in East Heilongjiang, China
Authors zhao J , Fu Y, Wang Y, Bao M, Li H, Qiu H
Received 28 April 2022
Accepted for publication 3 August 2022
Published 16 August 2022 Volume 2022:15 Pages 4549—4555
DOI https://doi.org/10.2147/IDR.S372703
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Jing zhao,1,2,* Yu Fu,3,* Yong Wang,1 Mingjia Bao,3 Huiling Li,1 Hongbin Qiu2
1Department of Microbiology, The First Affiliated Hospital of Jiamusi University, Jiamusi, People’s Republic of China; 2Key Laboratory of Microecology-immune Regulatory Network and Related Diseases School of Basic Medicine, Jiamusi University, Jiamusi, People’s Republic of China; 3Department of Microbiology, Center for Disease Control and Prevention of Jiamusi, Jiamusi, People’s Republic of China
*These authors contributed equally to this work
Correspondence:Hongbin Qiu, Key Laboratory of Microecology-immune Regulatory Network and Related Diseases School of Basic Medicine, Jiamusi University, Jiamusi, People’s Republic of China, Email [email protected]
Objective: To investigate drug resistance and homology of pathogens that can cause infective diarrhea in East Heilongjiang, China, and provide suitable references for the clinical treatment.
Methods: The process of isolation and identification of pathogenic bacteria in the faeces and blood of diarrhea patients was conducted. The drug resistance of the isolated pathogenic bacteria was analyzed by VITEK 2 compact drug sensitivity analysis system, and the strain homology was determined by pulsed field gel electrophoresis (PFGE).
Results: From 2014 to 2018, 28 different strains of pathogenic bacteria were isolated from 657 samples of faeces and blood, including 23 strains of Salmonella, 3 strains of Vibrio parahaemolyticus and 2 strains of diarrheal Escherichia coli, with detection rates of 3.5%, 0.46% and 0.30% respectively. Among them, 22 strains of Salmonella exhibited significant drug resistance, with a drug resistance rate of 95.65%. A total of 11 different PFGE fingerprints of 23 Salmonella strains were also obtained, with a homology of 56.5– 100%.
Conclusion: Among the various pathogens causing infective diarrhea in East Heilongjiang, China, Salmonella displayed multiple drug resistance and its distribution was found to be polymorphic.
Keywords: diarrhea, pathogen, drug resistance, homology
Introduction
There are several kinds of bacteria, which can cause infectious diarrhea. Salmonella is one of the most common intestinal pathogenic bacteria. The disease outbreak and hospitalization caused by Salmonella are a major source of bacterial infection. Salmonella can cause illness in both human as well as animals and can spread through contact or food chain.1 Worldwide, Salmonella causes about 115 million diseases and three hundred and seventy thousand deaths every year. In China, 70–80% of are caused by Salmonella infection, which is significantly higher as compared to other pathogenic bacteria. In Africa, the number of deaths caused by Salmonella infection reaches millions every year;2 in Europe, the annual bacterial infection rate caused by Salmonella is 0.035%,3 and in United States of America, the annual incidence rate of Salmonella infection is 15.4%.4 A number of antibiotics are used as the main drugs for the treatment of diarrhoeal diseases caused by pathogenic bacteria. For example, quinolones are mostly used to treat Salmonella infection in adults, whereas cephalosporins are the first choice for Salmonella infection in children.5,6 However, due to the unreasonable use of antibiotics, many drug-resistant pathogenic bacteria have emerged and mutated into dominant bacteria,7 thereby resulting in the loss of some antibiotics in clinical use. At present, the phenomenon of bacterial drug resistance is very common in China and multi drug resistance and even pan drug resistance strains continue to appear, which serve as serious threat to public health and safety. The widespread presence of drug-resistant Salmonella has become a worldwide public health concern.8 In this article, the drug resistance mechanisms of Salmonella was studied to provide a theoretical basis for developing suitable medications against infectious diarrhea, reducing suffering of the patients and improving the treatment efficiency.
Materials and Methods
Reagents
The gram-negative aerobic bacteria susceptibility test plate was obtained from Shanghai XingBai Biotechnology Co., Ltd. (China) and Thermo Fisher Scientific (U.S.A). Protease K (Merck, U.S.A) SeakemGold Agarose (Lonza, U.S.A) and Restriction Enzyme Xba I (New England Biolabs, U.S.A) were also used for the various experiments.
Materials
Faecal and blood samples (657) were collected from diarrhea patients in a hospital in East Heilongjiang, China. We obtained oral consent from the patients. The study protocol was approved by the Ethics Committee of Jiamusi University Clinical Medical College (approval No. 0378).
Method
Isolation and Identification of the Pathogenic Bacteria
After isolation and culture of the collected samples, the suspected Salmonella colonies were selected for pure culture, and the pure cultures were identified by VITEK 2 compact (bioMerieux, France). The serotype was thereafter determined by serum agglutination test on the identified Salmonella strain.
Drug Resistance Analysis
VITEK 2 compact was used to analyze the drug sensitivity of Salmonella. The specific operation steps used were as follows. Salmonella and quality control standard strains Escherichia coli (ATCC 25922) and Klebsiella pneumoniae (ATCC 700603) were first purely cultured overnight to prepare 0.5 McFarland value bacterial suspension. The drug sensitive plate and bacterial suspension were then arranged on the shelf and identified. The results were thereafter recorded. First, Salmonella was cultured overnight and the bacterial suspension was diluted to 0.5 McFarland value with 0.45% saline. The drug sensitivity card was filled as well as sealed and then loaded on to the instrument incubator. The instrument was monitored for the bacterial growth in each well within the specified time. After the incubation cycle, the mic value (or test result) of each antibacterial agent on the drug sensitivity card was obtained.)
PFGE Detection
Referring to the “pulse field gel electrophoresis (PFGE) operation standard”, Salmonella pulse field gel electrophoresis standard operation protocol was employed to carry out the homology analysis for Salmonella.
Purely cultured Salmonella and standard strain H9812 were incubated into 400 μL bacterial suspension with CSB (cell suspension buffer) respectively, with a concentration of 4.0–4.5 Michaelis units, and 400 μL of 1% Seakem Gold were used to make the small gel blocks. The prepared gel blocks were placed into 5 mL CLB (Cell Lysis Buffer) and incubate in a 54°C water bath shaker for 2 hours at a rotational speed of 130 r/min by HZS-HA water bath cradle (Harbin East Union Electronic Technology Development Company, China). Clean the cracked rubber block twice with sterile ultra pure water, and then washed four times with TE (Tris EDTA Buffer). Thereafter, the gel blocks were put into Xba I. for enzyme digestion to ensure that the gel blocks were immersed under the liquid level and further incubated in a 37 ion to ensure that the gel blocks were ter enzyme digestion were stuck on the comb teeth and converted into electrophoretic glue with 1% Seakem Gold glue. The electrophoretic conditions used were as follows. The initial and final conversion times were 6.76 and 35.38 s by CHEF Mapper pulsed field gel electrophoresis system and gel imaging system (Bio-Rad, U.S.A), respectively, and the electrophoresis time was 18 h. After electrophoresis, the gel was placed in 400 mL Gelred diluent and stained for 30 min, and then decolorized using DI (deionized water) for 1 h. The gel imaging system (Bio-Rad, U.S.A) and BioNumerics version 7.6.3 software (Applied Maths, Belgium) were used to analyze the electrophoresis results.
The criteria used for analysis of results of pulsed field gel electrophoresis was based on the method recommended by Tenover et al. In brief, those with similar band size and number in the PFGE map were identified as the same type, but those with 3 or less bands were identified as the different subtypes of the same type. In addition, those with more than 3 bands were identified as the different types. The genes of the different subtypes of the strains of the same type were correlated, which were derived from the same parent, but the strains of the different types had no epidemiological correlation.9
Statistical Method
All the data was statistically analyzed by chi-square test using SPSS 22.0 software. Significance threshold was set at p-value of ≤0.05.
Results and Analysis
Contamination of the Pathogenic Bacteria in Faeces and Blood of Diarrhea Patients in East Heilongjiang, China
Among the 657 faeces and blood samples from patients, 28 strains of different pathogenic bacteria were isolated, including 23 strains of Salmonella, 3 strains of Vibrio parahaemolyticus and 2 strains of diarrheal Escherichia coli, with prevalence rates of 3.5%, 0.46% and 0.30%, respectively. Additionally, 8 distinct strains of pathogenic bacteria were obtained from blood, accounting for 28.57%, and 20 strains were from faeces, accounting for 71.43%. It was observed that the pathogenic bacteria were mainly isolated from faeces. There are 7 diverse serotypes of 23 Salmonella strains by detached, and Salmonella Enteritidis is the main serotype. The details have been shown in Table 1.
 |
Table 1 Serotype of Salmonella |
Drug Resistance of the Different Pathogens
The results of drug sensitivity assay showed that 22 out of 23 Salmonella strains (accounting for 95.65%, 22/23) exhibited drug resistance against 13 different antibiotics, and Nalidixic acid showed the highest drug resistance rate (56.52%), but they were sensitive to ceftazidime and amikacin. The details have been shown in Table 2 below.
 |
Table 2 Drug Sensitivity of Salmonella |
Among the 22 drug resistance Salmonella strains, 3 strains were single drug resistance (13.04%, 3/23) and 19 strains were multi drug resistance (82.61%, 19/23), thereby resulting in a total of 16 drug resistant spectra. Interestingly, the number of the strains with double drug resistance was the largest, accounting for 34.78% (8/23). The details have been shown in Table 3.
 |
Table 3 Drug Resistant Spectrum of Salmonella |
Pathogen Homology
PFGE molecular typing was conducted for 22 different Salmonella strain that were drug resistant after Xba I enzyme digestion. A total of 11 distinct fingerprints were obtained, and the cluster recognition degree was found to be 56.5–100%. The identification degree of the clusters with higher fingerprint similarity was 92.9–100%. The homology between the strains SMJMS13, SMJMS17 and SMJMS16 was observed to be 100%, and these were all isolated from faeces. The homology of SMJMS02, SMJMS03, SMJMS05, SMJMS11, SMJMS14, SMJMS18, SMJMS19 and SMJMS20 was found to be 100%, which were all isolated from faeces. The homology between SMJMS09 and SMJMS10 were 100%, which were all obtained from faeces. The homology between SMJMS06 and SMJMS12 were 100%, which were isolated from blood and faeces respectively. The degree of recognition between these strains was 91.5–96.0%. For the strains with higher cluster similarity in other clusters, the homology between SMJMS01 and SMJMS21 was 92.9%, which were obtained from blood and faeces respectively, as shown in Figure 1.
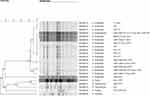 |
Figure 1 PFGE fingerprint, serotype and drug resistance spectrum of 22 Salmonella strains. Note: No drug resistance, no drug resistance spectrum. |
Discussion
Twenty-three strains of Salmonella were isolated from 657 faeces and blood samples, which was significantly higher than other pathogenic bacteria, but slightly different from the statistical results of national foodborne diseases in 2019. The number of patients affected with the microbial factors was the highest, accounting for 52.01% (12,738/24,491), among which Vibrio parahaemolyticus (30.25% (3853/12,738)) and Salmonella (28.44% (3623/12,738)) were the main pathogenic bacteria, followed by Staphylococcus aureus (8.03% (1023/12,738)) Bacillus cereus (6.27% (799/12,738)) and diarrhoeal Escherichia coli (4.22% (537/12,738)).10 In this study, 23 different strains of Salmonella were isolated from 657 faeces and blood, among which 8 strains were obtained from blood and 15 strains were from faeces. Among them, 22 strains had drug resistance, of which the drug resistance rate against nalidixic acid was the highest, reaching 56.52%. This finding indicated that over the years, the frequency and dose of quinolone antibiotics have been relatively high, thereby resulting in the alteration of drug resistance of Salmonella against the antibiotics, and the increase of the resistance of these strains to nalidixic acid, which is one of the primary reasons for the significant diarrheal symptoms of patients. It might be also possible that the combination of virulence genes and other factors can enhance the virulence of the strains, thus rendering it easier for the contacted person to get sick. Thus, it is necessary to further analyze and study the virulence genes and other influencing factors. When using antibiotics for the treatment of diarrhea disease, effective antibiotics should be selected according to the drug resistance of local faeces and blood strains. For instance, Salmonella in Xinxiang City, Henan Province was found to be most sensitive to cephalothin and gentamicin.11 Salmonella in Guangzhou City was sensitive to ceftriaxone, cefepime, imipenem, meropenem, cefoperazone/sulbactam and amikacin.12 Salmonella found in Urumqi City was sensitive to ciprofloxacin, ceftriaxone, gentamicin, kanamycin, cefoxitin and amikacin.13 This may be related to the potential use of distinct classes of antibiotics in the different regions. The Salmonella strains isolated in Urumqi had the highest sensitivity rate (up to 100%) towards ceftazidime and amikacin. According to the drug resistance of Salmonella strains infected by diarrhea patients in Urumqi City, antibiotics can be selected to achieve better clinical treatment outcome. Overall, understanding the source of Salmonella infection is of great significance to control the occurrence and development of various diseases.
Further analysis of Salmonella homology, serotype and drug resistance revealed that among the four diverse groups of strains with the same homology, the drug resistant spectra of the three groups of strains were different. Among all, only three strains SMJMS18, SMJMS19 and SMJMS20 drug resistant spectra in one group was the same, but in the other five strains were different, indicating that majority of the strains with the same PFGE band type were different from most drug resistant spectra, which was consistent with the previously reported studies.14 There was minimal difference in PFGE bands among the different strains with the same Salmonella serotype. The fingerprint identification degree of 17 strains of Salmonella enteritidis in this study ranged between 59.1% and 100%, showing six distinct bands, which was different from those obtained from other regions. It was observed that the relationship between the genetic relation of the strains and the interaction of drug resistance and serotype remains unclear and thus further research is needed.
The fingerprints, sources, serotype and drug resistant spectra of the three strains SMJMS18, SMJMS19 and SMJMS20 isolated from the faeces of the diarrhea patients were found to be consistent. It can be determined that they arise from the same strain, and it can be inferred that the three patients might have been exposed to the same pollution source, thereby resulting in the consistent testing results of Salmonella strains isolated from the faeces of the three patients. However, epidemiological investigation is needed to further determine the identical pollution source and supplement relevant information to complete the pathogen tracing process. Overall, 11 PFGE fingerprints of other 19 strains of Salmonella were obtained, thus indicating polymorphic distribution, which was similar to the situation in some other regions.15,16 The findings of this study indicate that a thorough investigation of the homology and drug resistant spectrum of diarrhea pathogens can provide optimal references for an effective treatment of the clinical diarrhea patients and a strong theoretical support for developing clinical guidance for medication.
In this study, the drug resistance and homology of Salmonella isolated from the blood or faeces of the diarrhea patients were analyzed for the first time to clarify the mechanisms of drug resistance in response to Salmonella infection. The findings generated can effectively guide the clinicians to accurately select the type of antibiotics to be administered and provide targeted diagnostic basis, to reduce the duration of treatment and effectively treat infected patients. In addition, we should also increase health publicity, improve people’s awareness, prevent possible harm, block the possible transmission path of the pathogenic bacteria, reduce the harmful effects of pathogenic bacteria on the public health, and effectively prevent the occurrence of diarrhoeal diseases.
Conclusions
Salmonella isolated from this city ranged from both single drug resistance to eight-fold drug resistance, and most of which was concentrated in two-fold or more, which was similar to the drug resistance situation of these strains in some other regions of China. These findings indicated that the multiple drug resistance of Salmonella in our city was significant, which substantially increased the difficulty of the treatment of diarrhoeal patients. In order to reduce the production of multi drug resistant Salmonella strains, it is recommended that antibiotics should be used reasonably for the treatment of diarrhea diseases. Additionally, the mechanisms of drug resistance should be thoroughly investigated to achieve effective clinical treatment against diarrhea disease.
Data Sharing Statement
The data used to support the findings of this study are included within the article.
Ethics Approval and Consent to Participate
All the protocols were approved by the Ethics Committee of Jiamusi University Clinical Medical College (0378). Patient informed consent was waived as the samples were collected from existing specimens retrieved during routine procedures and did not require further involvement from patients. For patients who were actively screened, we obtained oral consent from the patients.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, drafting and revising the article, gave final approval of the version to be published, have agreed on the journal to which the article will be submitted and agreed to be accountable for all aspects of the work. The final manuscript was reviewed and approved by all authors.
Funding
Health commission scientific research plan projects of Heilongjiang (2020-356).
Disclosure
The authors have no competing interests.
References
1. Arora D, Kumar S, Jindal N, et al. Prevalence and epidemiology of Salmonella enterica serovar Gallinarum from poultry in some parts of Haryana. Vet World. 2015;8(11):1300–1304. doi:10.14202/vetworld.2015.1300-1304
2. Cabello FC. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol. 2006;8(7):1137–1144. doi:10.1111/j.1462-2920.2006.01054
3. McGuinness S, McCabe E, O’Regan E, et al. Development and validation of a rapid real-time PCR based method for the specific detection of Salmonella on fresh meat. Meat Sci. 2009;83(3):555–562. doi:10.1016/j.meatsci.2009.07.004
4. Marder EP, Cieslak PR, Cronquist AB, et al. Incidence and trends of infections with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance: foodborne diseases active surveillance network, 10 US Sites, 2013–2016. Morb Mortal Wkly Rep. 2017;66(15):397–403. doi:10.15585/mmwr.mm6615a1
5. Cheng MW, Lee CM, Wang NY, et al. Clinical characteristics in adult patients with Salmonella bacteremia and analysis of ciprofloxacin-nonsusceptible isolates. J Microbiol, Immunol Infect. 2015;48(6):692–698. doi:10.1016/j.jmii.2015.09.001
6. Britto CD, Wong VK, Dougan G, et al. A systematic review of antimicrobial resistance in Salmonella enterica serovar Typhi, the etiological agent of typhoid. PLoS Negl Trop Dis. 2018;12(10):e0006779. doi:10.1371/journal.pntd.0006779
7. Clemente L, Manageiro V, Jones-Dias D, et al. Antimicrobial susceptibility and oxymino-β-lactam resistance mechanisms in Salmonella enterica and Escherichia coli isolates from different animal sources. Res Microbiol. 2015;166(7):574–583. doi:10.1016/j.resmic.2015.05.007
8. Eddra A, Filali FR, Karraouan B, et al. Prevalence, molecular and antimicrobial resistance of Salmonella isolated from sausages in Meknes. Microb Pathog. 2017;105:340–345. doi:10.1016/j.micpath.2017.02.042
9. Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi:10.1128/jcm.33.9.2233-2239.1995
10. Li H, Guo Y, Song Z, et al. Analysis of surveillance data on foodborne disease outbreaks in Chinese Mainland in 2019. Chin J Food Hygiene. 2021;33(06):650–656. doi:10.13590/j.cjfh.2021.06.003
11. Liu G, Yu T. Study on antimicrobial resistance and plasmid-mediated quinolone resistance of foodborne Salmonella isolates. Biotechnol Bulletin. 2014;8:202–207. doi:10.13560/j.cnki.biotech.bull.1985.2014.08.034
12. Zhou R, Li L, Su J, Li B, Xu Z. Detection of class I integron and analysis of drug resistance gene cassette of foodborne Salmonella. Sci Technol Food Industry. 2014;35(01):
13. Ge K, Wu Y, Yang B, et al. Quinolone resistance and related gene analysis of Salmonella in beef and mutton retailed in Urumqi. Food Sci. 2017;38(04):107–112.
14. Lin J, Qin C, Lai J, Shu G, Wu C. Detection and analysis of plasmid-mediated quinolone resistance genes in Salmonella isolates from food animals. Acta Veterinaria Et Zootechnica Sinica. 2012;43(05):803–809.
15. Yang B, Zhang X, Qu D, et al. Serotypic and genotypic characterization of Salmonella Serovars from retails meat in Shaanxi province (2007–2008). Acta Microbiologica Sinica. 2010;50(05):654–660. doi:10.13343/j.cnki.wsxb.2010.05.018
16. Li M, Zhang X, Qiang L, et al. Epidemiologic and etiologic characteristics of foodborne diseases active surveillance in Baiyin City, 2016–2018. Chin J Food Hygiene. 2020;32(01):72–76. doi:10.13590/j.cjfh.2020.01.014
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
