Back to Journals » Cancer Management and Research » Volume 11
DR7 encoded by human herpesvirus 6 promotes glioma development and progression
Authors Gu B, Li M , Zhang Y, Li L, Yao K, Wang S
Received 10 July 2018
Accepted for publication 10 January 2019
Published 12 March 2019 Volume 2019:11 Pages 2109—2118
DOI https://doi.org/10.2147/CMAR.S179762
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Beicheng Sun
Bin Gu,1 Meng Li,2 Yan Zhang,3 Lingyun Li,4 Kun Yao,5 Shizhi Wang3
1Department of Neurosurgery, Zhongda Hospital, School of Medicine, Southeast University, Nanjing, China; 2Department of Neurosurgery, Suqian First Hospital, Suqian, China; 3Key Laboratory of Environmental Medicine Engineering, Ministry of Education, School of Public Health, Southeast University, Nanjing, China; 4Department of Microbiology and Immunology, Nanjing Medical University, Nanjing, China; 5Department of Developmental Genetics, Nanjing Medical University, Nanjing, China
Purpose: We previously identified human herpesvirus 6 (HHV-6) infection in the pathogenesis of glioma. Direct repeat (DR)7, encoded by HHV-6, has been reported to possess malignant transforming activity and involved in Hodgkin’s lymphoma carcinogenesis. Here, we aimed to determine the role of DR7 in the development and progression of glioma.
Patients and methods: A total of 27 glioma and 30 normal brain tissues were collected for detection of DR7. Glioma cell proliferation, colony formation, cell cycle, migration, invasion and angiogenesis were evaluated by Cell Counting Kit-8 (CCK-8), soft agar, propidium iodide staining, wound healing, Transwell and chick embryo chorioallantoic membrane assays, respectively. The potential mRNA targets of DR7 were determined using mRNA microarray and validated via Western blot and ELISA.
Results: DR7 could be detected in the 13 glioma tissues with a positive rate of 48.15%, but only the 5 normal brain tissues with a lower positive rate of 16.7%. The two strains of cells isolated from glioma tissues were also found to express DR7. CCK-8 and soft agar assays showed enhanced proliferation and colony formation in the cells expressing DR7 which might be in relation to acceleration of the G1/S phase transition by DR7. Further analyses showed that DR7 could promote glioma cell migration, invasion and angiogenesis. Expression profiles identified hundreds of differentially expressed mRNAs, among which P53, extracellular matrix (ECM) fibronectin, integrin receptor ITGβ5 and specific inhibitors of MMPs, tissue inhibitor of MMPs (TIMP)-2 and TIMP-4, were downregulated, whereas ECM-degrading proteinase MMP-3, proinflammatory cytokines IL-1β, IL-6 and IL-8, were upregulated by DR7, respectively.
Conclusion: We observed existence of DR7 in the glioma tissues, and overexpression of DR7 could promote glioma cell development and progression, which might be through creating an inflammatory microenvironment and enhancing degradation of ECM.
Keywords: HHV-6, DR7, development, progression, glioma
Introduction
Glioma is a common malignancy in human brain tumors and its incidence is about 5 cases per 100,000 people.1 There are over 140,000 new patients in the USA each year. In addition, about 13,000 people die each year because of this related disease.2 At present, the pathology of glioma is still unclear. Some scholars believe that the occurrence and development of tumors can be promoted by intrinsic factors and external factors. Intrinsic factors include activation of proto-oncogenes and local mutations in tumor suppressor genes. Environmental factors include chemical and physical factors, such as chemical carcinogens, biological factors and other reasons.3 Among them, the research on the role of viruses in the development of glioma has received increasing attention.
Human herpesvirus 6 (HHV-6) is one of the most widely distributed linear double-stranded DNA viruses.4 In 1986, HHV-6 was isolated for the first time.5 Later studies found two distinct variants, named as HHV-6A and HHV-6B.6 Although the genomes of HHV-6A and HHV-6B are colinear and shared an overall identity of 90%, the two groups showed distinct epidemiology and disease associations, biological and immunological properties, and in vitro tropism for selected T-cell lines.7 For example, HHV-6B caused 97%–100% of the primary infections by these viruses and the infections mostly occur between the ages of 6 and 12 months.8 Research on the epidemiology of HHV-6A infection is less, and one report has indicated that HHV-6A infection is acquired later in life and the primary infection is typically without clinical symptoms.9
HHV-6 has the characteristics of transformation, transactivation and carcinogenesis,10 and many diseases of nervous systems are associated with HHV-6, such as encephalitis,11 multiple sclerosis and glioma.12 HHV-6 has a unique region (U) of 143–145 kb, flanked by 8–9 kb of terminal direct repeats (DRs). The open reading frames (ORFs) of DR are designated as DR1–DR7,9 among which 357 amino acids in the SalI-L fragment are ORF-1, also named as DR7.13 The length of DR7 is 1,092 bp, and its protein can be detected after 18 hours of virus infection, but it is not expressed during viral latency. DR7 can transform NIH3T3 cells in vitro and form tumors in nude mice.13 Further study shows that DR7 can bind to p53 and lead to impaired p53 protein function, suggesting that DR7 is one of the key tumor genes of HHV-6.14 DR7 locates at positions 5,629–6,720 of the HHV-6 genome, which partially overlaps with spliced DR6 at positions 4,725–5,028 and 5,837–6,720. It was reported15 that the homologous gene in HHV-6B, that is, DR6B, encodes a nuclear protein which can interact with the viral DNA processivity factor p41 rather than p53. Borenstein et al16 cloned the intact HHV-6A genome into bacterial artificial chromosome (BAC) vectors and found HHV-6A BACs and their parental DNAs to contain short 2.7 kb DRs. Further studies revealed that the deletion spans positions 60–5,545 in DRL (left DR), including genes encoded by DR1 through the first exon of DR6. The conserved pac-2–pac-1 packaging signals, the DR7 ORF and the DR6 second exon were not deleted. Thus, the biological role of DR7 is different from DR6, regardless of overlapping sequences.
We previously revealed involvement of HHV-6 in the pathogenesis of glioma. We detected higher percentages of HHV-6 DNA and protein in the tissues of glioma than in the tissues of normal brain, which was in accordance with Crawford et al’s3 findings, and directly isolated a strain of HHV-6A from glioma cyst fluid. In addition, we detected high levels of IL-6, IL-8, tumor necrosis factor (TNF)-α and TGF-β in the cyst fluid specimens from HHV-6-positive patients with glioma, and also revealed that HHV-6A infection could promote production of IL-6, IL-8 and TGF-β in astrocyte cultures. We proposed that proinflammatory cytokines induced by HHV-6 infection might provide a chronic inflammatory environment which facilitates the development of glioma.17 At present, there are few reports about the relationship between DR7 and glioma. In this study, we aimed to determine whether DR7 contributes to glioma development and progression.
Patients and methods
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional review board (IRB) of Southeast University and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All applicable international, national and/or institutional guidelines for the care and use of animals were followed. The study was approved by the IRB of Nanjing Medical University and Southeast University.
Patients and samples
A total of 27 glioma and 30 normal brain tissues were collected for analysis of DR7 expression. The glioma specimens were obtained from patients with glioma undergoing surgery in the First Affiliated Hospital of Nanjing Medical University from 2008 to 2013. The normal specimens were obtained from patients with cerebral hemorrhage or brain trauma undergoing surgery in the same hospital during that time. The patients whose tissues were used provided written informed consent.
Cell culture
Human glioma cell lines U87 and U251 were obtained from Prof Yao (Department of Microbiology and Immunology, Nanjing Medical University). U87 and U251 cells were cultured in DMEM containing 10% FBS and placed in a 37°C, 5% CO2 cell incubator. The use of the cell lines was approved by the IRB of Southeast University.
Lentivirus vector construction and packaging
The pMD18-T-DR7 plasmid carrying the DR7 gene (HHV-6A U1102 strain) was maintained by the Laboratory of Viral Immunology, Department of Microbiology and Immunology, Nanjing Medical University. The lentivirus vector pLenti6.3-MCS- IRES2-EGFP carrying the EGFP gene, the packaging plasmids pLP1, pLP2, pLP/VSVG, and the competent cell Stbl3 were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Primers used for amplifying the DR7 gene are listed as follows: P1: 5′-GACACTTAATTAAGCCGCCACCATGCGGCATCTCCCGTTC CACGGGATGC-3′; P2: 5′-GTGTCGCTAGCTTAGTGATGATGATGATGATGCT TCACCTCCGTGGCGTCTAATCC-3′. P1 and P2 contained PacI and NheI restriction sites, respectively.
Lentiviral packaging was performed using Stbl3 cell, and virus titers were determined by counting the cells expressing EGFP. The lentiviral expression vector was successfully packaged and the virus titer was 108 TU/mL. After lentivirus infection of DR7, U87 cells were subjected to flow cytometry and blasticidin screening until they stably expressed DR7.
RNA isolation and mRNA microarray
Total RNA of U87-NC-EGFP and U87-DR7-EGFP cells was extracted using TRIzol reagent (Thermo Fisher Scientific). Global gene expression was measured with the Affymetrix HG-U133 Plus 2.0 chip. A total of 10 µg of RNA was used for synthesize double-stranded cDNA using the One-Cycle cDNA Synthesis Kit (Affymetrix). The resulting cDNA was used as a template for in vitro transcription (IVT) of CRNA using IVT Labeling Kit (Affymetrix). A hybridization cocktail containing 10 µg of CRNA was hybridized to U133 Plus 2.0 GeneChips (Affymetrix) for 16 hours at 45°C in a rotating oven. Finally, the chips were washed, stained and scanned on a high-resolution scanner following the protocol. A twofold cutoff difference was applied to select the up- and downregulated mRNAs.
Cell Counting Kit-8 (CCK-8) testing
Cells in logarithmic phase of growth were added to the 96-well plates (100 µL/well; about 1×104). CCK-8 was detected at 24, 48 and 72 hours of culture. The spectrophotometric plate reader was used to measure the OD of each well at a wavelength of 450 nm.
Soft agar colony formation assay
The mixture containing 0.6% agarose and 2× DMEM (containing 2% antibiotics and 20% FBS) in a 1:1 ratio was taken in sterile tubes. Cells with a density of 1×105/mL were suspended in the tubes and then poured onto 1 mL of a 1.2% agar medium base layer in six-well plates. After 10–14 days of culture, the cells were placed under an inverted microscope and the number of colonies (>1 mm) was observed.
Cell scratch assay
Cells in logarithmic phase of growth were seeded at 2×105 cells/mL in six-well plates and cultured for 24 hours. A sterile 100 µL pipette tip was used to make a cell-free wound. Cells were photographed at 6, 12 and 24 hours, and the width of the scratches was measured.
Transwell invasion assay
The invasion assay was performed using Transwell chambers (Corning Life Sciences, Tewksbury, MA, USA) with a pore size of 8 µm. A total of 100 µL cells at a density of 1×106/mL were added to the upper compartment of invasion chambers and 500 µL of DMEM supplemented with 20% FBS was added to the lower chamber. After 24 hours of incubation at 37°C, the migrating cells were fixed with formaldehyde and stained with 0.1% crystal violet and counted under a microscope.
Chick embryo chorioallantoic membrane (CAM) assay
Fertilized chicken eggs were used after incubating them at 37°C, 5% CO2 for 4 days. To expose the CAM, a 1×1 cm window was made in the shell. Eggs were randomly divided into three groups with six eggs per group. The cells of U87, U87-NC-EGFP and U87-DR7-EGFP premixed with ice-cold Matrigel were placed on the CAM, respectively. Ten days later, the CAMs with tumors were cut and fixed with 4% paraformaldehyde. The vessel number from three random fields around the tumors was pictured using a microscope.
Cell cycle detection
Cell cycle was determined using propidium iodide (PI) staining and measured by flow cytometry as previously described.18 Briefly, cells were washed with PBS, fixed with 70% ethanol, resuspended in PBS containing PI/RNase and analyzed by flow cytometry.
Quantitative PCR (qPCR)
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific) and subsequently reverse transcribed with M-MLV reverse transcriptase (Toyobo, Osaka, Japan) at 42°C. For real-time qPCR, cDNA fragments were amplified using SYBR Green Real-time PCR Master Mix (Toyobo) according to the manufacturer’s instructions. qPCR primers of DR7 are listed as follows: ACTTTAAGAGGATC CCGCCACCATGAGGGCG (forward) and GCAGCCGGATCCACTAGTAACG GCCG (reverse).
ELISA
Fibronectin (FN), IL-1β, IL-6, IL-8, tissue inhibitor of MMPs (TIMP)-2 and TIMP-4 in the U87 cell culture supernatants were determined using ELISA kits (Bender, Vienna, Austria) as previously described.17 All ELISA assays were performed in triplicate.
Western blot
Cells were lysed using a detergent lysis buffer (50 mM Tris, pH 7.4; 150 mM NaCl; 1% NP-40; 0.5% sodium deoxycholate; 0.1% SDS and 1% phosphatase inhibitor cocktail [Sigma-Aldrich, St Louis, MO, USA]). Western blots were performed as previously reported19 using antibodies of P53, TIMP-2, TIMP-4, ITGβ5 and MMP-3 (CST, Danvers, MA, USA). β-Actin was used as an internal control (Bioworld, Shanghai, China).
Immunofluorescence (IF) and immunohistochemistry (IHC) assays
The primary cultured human glioma cells were fixed in 4% paraformaldehyde and stained with DR7 antibody (provided by the HHV6 Foundation) followed by a secondary antibody labeled with Rhodamine, and GFAP antibody (a prototypical marker for astrocytes; CST) followed by a secondary antibody labeled with fluorescein isothiocyanate. IHC was performed on paraffin-embedded sections. Series of sections of glioma specimens were incubated with DR7 antibody followed by incubation with 3,3′-diaminobenzidine (Zhongshan Biotech, Guangdong Sheng, China) to produce brown precipitates.
Statistical analysis
The experiments were repeated three times independently. The data were expressed as mean ± SD. Comparison of categorized and continuous variables between groups was performed using chi-squared test and ANOVA, respectively. SPSS 13.0 was used for statistical analysis, and P<0.05 was considered statistically significant.
Results
Expression of DR7 in glioma tissues
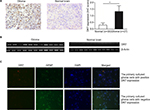
To determine whether HHV-6 expresses DR7 during glioma development, a total of 27 glioma and 30 normal brain tissues were collected for IHC analysis of DR7. The results showed DR7 was mainly expressed in the cytoplasm (Figure 1A) and there were 13 glioma tissues having positive DR7 staining, with a positive rate of 48.15%. However, there were only five normal brain tissues having DR7 expression with the positive rate being 16.7%. The DR7 expression was higher in the glioma tissues than in the normal brain tissues and the difference reached statistical significance. To verify the expression of DR7, we also tested the transcripts of DR7 in the eleven glioma tissues with positive IHC staining of DR7 and eleven normal brain tissues with negative IHC staining of DR7. The reverse transcriptase-PCR (RT-PCR) assay (Figure 1B) displayed that all the eleven glioma tissues had positive DR7 expression while all the eleven normal brain tissues had negative DR7 expression, in line with the IHC findings. To further validate the existence of DR7 in the development of glioma, we isolated two strains of primary human glioma cells for detection of DR7. As shown in Figure 1C, DR7 could be detected by IF assay. Taken together, our results suggested that DR7 is involved in the carcinogenesis of glioma.
DR7 favors cell proliferation and colony formation
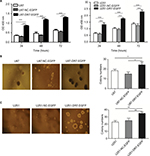
As shown in Figure 2A, U87AND U251 cells overexpressing DR7 had the highest proliferation rate than the blank and negative control (NC)-treated U87 and U251 cells, respectively, on assessing using CCK-8 assay. To determine the effect of DR7 on the colony-forming ability of glioma cells, soft agar colony cultures of the three groups of cells were performed. After 2 weeks, the number of colonies was 24.33±3.06, 15.00±2.00 and 18.33±1.53 for the U87-DR7-EGFP, U87-NC-EGFP and U87 cells, respectively (Figure 2B). The U87-DR7-EGFP cells had the highest colony number than the two other control cells. Repeating the soft agar assay in the U251 cells showed similar results (Figure 2C).
DR7 promotes cell cycle progression
To determine whether DR7 enhances cell proliferation and colony formation through promoting cell cycle progression, we tested the cell cycle distribution of the three groups of cells by flow cytometry. The percentage of G0/G1, S and G2/M phases of cell cycle for different cell groups is shown in Figure 3A. The highest percentage of S and G2/M phases and the lowest percentage of G0/G1 phase were seen in U87-DR7-EGFP cells among the three groups of cells (Figure 3B). The results indicated that DR7 promotes the transition of cells from G1 to S phase, which facilitates cell proliferation. Repeating the cell cycle assay in the U251 cells showed similar results (Figure 3C, D), although the differences in the percentage of G2/M phase among the U251-DR7-EGFP, U251-UC-EGFP and U251 cells did not reach statistical significance.
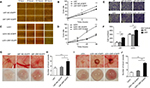
DR7 enhances cell migration, invasion and angiogenesis of glioma
Cell scratch assays demonstrated that the U87 and U251 cells transfected with DR7 were significantly healed after 6 hours of culture compared with their respective control cells. After 12 hours of culture, the difference in healing rates among the three groups of cells was even more significant (Figure 4A–D). In addition, Transwell invasion assays revealed the highest number of invading cells in relation to U87 and U251 cells transfected with DR7 (Figure 4E, F). Tumor growth and metastasis needs enhanced angiogenesis to supply extra oxygen and nutrients.20,21 We also evaluated whether DR7 could regulate angiogenesis of glioma cells. CAM assays displayed the highest vascular density for U87/U251-DR7-EGFP cells compared with their respective control cells (Figure 4G–J).
Identification and validation of targets of DR7 in glioma
We further applied mRNA microarray to identify the critical genes and pathways downstream of DR7 involved in glioma progression. There were 1,016 upregulated and 778 downregulated genes in the U87-DR7-EGFP cells compared with the control U87-NC-EGFP cells. Finally, we selected four upregulated genes, that is, IL-1β, IL-6, IL-1β and MMP-3, and five downregulated genes, that is, FN, P53, TIMP-2 and TIMP-4, and ITGβ5, for validation of their expression trend in response to overexpression of DR7 in U87 cells (Table 1). The selected genes are all well known as fundamental regulators in cell proliferation, migration, invasion and angiogenesis.
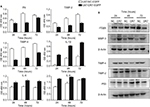
For the secreted proteins, such as FN, TIMP-2 and IL-8, we employed ELISA to determine their expression differences between the U87-DR7-EGFP cells and U87-NC-EGFP cells. As shown in Figure 5A, the expression of secreted FN, TIMP-2 and TIMP-4 was higher in the U87-DR7-EGFP cells than in U87-NC-EGFP cells, whereas the expression trend of IL-1β, IL-6 and IL-8 was just the opposite. We also tested the expression of ITGβ5, MMP-3 and P53 using Western blot. The results showed higher expression of MMP-3 and lower expression of ITGβ5 and P53 in the U87-DR7-EGFP cells than in U87-NC-EGFP cells. Moreover, the trend of differential expression was more pronounced over time (Figure 5B). TIMP-2 and TIMP-4 were also subjected to Western blot for further validation of their expression trend in response to DR7 overexpression, and they presented the same pattern of lower expression in the U87-DR7-EGFP cells (Figure 5B). Overall, the expression trend of all nine genes was in accordance with that of mRNA microarray.
Discussion
Recent studies suggest that the occurrence and development of glioma is closely related to HHV-6 infection.12,17 Kashanchi et al first found the SalI-L fragment of HHV-6 had the ability to transactivate HIV-1 long terminal repeat promoter and increase HIV-1 replication.22 Thereafter, they narrowed the SalI-L transforming region to the SalI-L-SH subfragment and found that ORF-1 (also designated as DR7), one of the ORFs within SalI-L-SH, could transform NIH3T3 cells. Furthermore, nude mice developed fibrosarcomas when they were injected with cells expressing DR7.13 PCR analysis of DNA sequences of DR7 identified its existence in some angioimmunoblastic lymphadenopathies, Hodgkin’s lymphoma (HL) and non-HL and glioblastomas.13 Consistent with Kashanchi, Lacroix et al also found DR7 could enhance the proliferation of Reed–Sternberg cells.10 Their findings suggest that DR7 has a tumorigenic property. We also observed existence of DR7 in the glioma tissues and two strains isolated from the primary glioma tissues. In addition, overexpression of DR7 could promote glioma U87 cells’ proliferation.
Angiogenesis can provide necessary oxygen and nutrients to and remove wastes from tumor cells, and is essential for tumor growth and metastasis.20,21 Caruso et al reported that HHV-6 infection could result in loss of angiogenic properties both in lymphatic endothelial cells (ECs) and in vascular ECs. They further identified U94/rep, a unique genomic characteristic of HHV-6, contributing to the antiangiogenic effect of HHV-6.23 Our unpublished data also suggested antiangiogenic characteristics for U94/rep in glioma. However, in the present study, we identified DR7, another gene of HHV-6, enhancing cell angiogenesis. Our result indicated that DR7 might play a proangiogenic role opposite to U94/rep in glioma. Therefore, increased angiogenic capacity in glioma cells infected with HHV-6 might be ascribed to overexpression of DR7, underexpression of U94/rep or both in combination.
So far, the underlying mechanisms of DR7 in human disease remain to be resolved. Kashanchi et al13 observed that DR7 could influence proliferation by binding to P53. Lacroix et al10 also observed in the HL that DR7 protein is bound to human P53 protein. In the present study, we found decreased expression of P53 in the glioma cells transfected with DR7. Whether there is a direct interaction between DR7 and P53 in glioma needs further investigation. Invasion and metastasis are two mechanisms for cancer spreading throughout the body. MMPs are a broad family of zinc-binding endopeptidases collectively degrading extracellular matrix (ECM) components, whose deregulation favors cancer cell invasion, metastasis and angiogenesis.24,25 The activated MMPs are inhibited by four mammalian TIMPs.26 The adhesive glycoprotein FN and integrin receptors play critical roles in tumor progression.27 MMP-3 (Stromelysin 1) is able to degrade FN, laminin, gelatins of type I, III, IV and so on. We observed in this study overexpression of MMP-3 and underexpression of TIMP-2, TIMP-4, FN and ITGβ5. The results indicated that DR7 could promote degradation of ECM which facilitates cells’ progression including migration, invasion and angiogenesis.
We previously found increased production of proinflammatory cytokines, such as IL-6, IL-8, TGF-β and TNF-α, in glioma cyst fluid specimens and astrocyte cultures.17 In this study, we also identified several cytokines, such as IL-1β, IL-6 and IL-8, deregulated in the glioma cells transfected with DR7. Thus, HHV-6 infection might create an inflammatory microenvironment that facilitates pathogenesis of glioma through DR7. In silico analysis showed that P53, IL-6, FN1 and ITGβ5 were all involved in PI3K-Akt signaling pathway (Table 1). It provides us a clue to determine the potential signal pathway which is regulated by DR7 to promote glioma progression. In addition, we also found NFKBIZ (NFKB Inhibitor Zeta) was upregulated 5.5-fold in the U87 cells overexpressing DR7. NFKBIZ could inhibit NF-kappa-B activity without affecting its nuclear translocation upon stimulation.28 In addition, CSF-3, LIF, IL6 and IL13RA2 were found to be upregulated 10.6-, 3.1-, 2.6- and 2.1-fold in the U87 cells overexpressing DR7, respectively. These genes are all involved in the JAK–STAT signaling pathway.29–32 NFκB and JAK–STAT pathways are found to be widely involved in glioma carcinogenesis.33,34 Thus, DR7 may promote glioma development and progression through the NFκB and JAK–STAT pathways besides the PI3K-Akt pathway.
Conclusion
DR7 could be detected in glioma with positive HHV-6 infection. Overexpression of DR7 was able to promote glioma proliferation, migration, invasion and angiogenesis, which might be through creating an inflammatory microenvironment and enhancing degradation of ECM.
Acknowledgments
We thank Dr Lukui Chen for his critical review of our manuscript and the HHV-6 Foundation for providing us DR7 antibody. This study was partly supported by National Natural Science Foundation of China (81872684, 81301698 and 81201520), Natural Science Foundation of Jiangsu Province (BK20171367 and BK20171489), Key Project supported by Medical Science and Technology Development Foundation, Jiangsu Provincial Health Commission (Z201409) and the Fundamental Research Funds for the Central Universities (2242018K40020).
Disclosure
The authors report no conflicts of interest in this work.
References
Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. | ||
Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789–794. | ||
Crawford JR, Santi MR, Cornelison R, Sallinen SL, Haapasalo H, MacDonald TJ. Detection of human herpesvirus-6 in adult central nervous system tumors: predominance of early and late viral antigens in glial tumors. J Neurooncol. 2009;95(1):49–60. | ||
Telford M, Navarro A, Santpere G. Whole genome diversity of inherited chromosomally integrated HHV-6 derived from healthy individuals of diverse geographic origin. Sci Rep. 2018;8(1):3472. | ||
Salahuddin SZ, Ablashi DV, Markham PD, et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234(4776):596–601. | ||
Ablashi D, Agut H, Berneman Z, et al. Human herpesvirus-6 strain groups: a nomenclature. Arch Virol. 1993;129(1-4):363–366. | ||
Iyengar S, Levine PH, Ablashi D, Neequaye J, Pearson GR. Sero-epidemiological investigations on human herpesvirus 6 (HHV-6) infections using a newly developed early antigen assay. Int J Cancer. 1991;49(4):551–557. | ||
Dewhurst S, McIntyre K, Schnabel K, Hall CB. Human herpesvirus 6 (HHV-6) variant B accounts for the majority of symptomatic primary HHV-6 infections in a population of U.S. infants. J Clin Microbiol. 1993;31(2):416–418. | ||
De Bolle L, Naesens L, De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev. 2005;18(1):217–245. | ||
Lacroix A, Collot-Teixeira S, Mardivirin L, et al. Involvement of human herpesvirus-6 variant B in classic Hodgkin’s lymphoma via DR7 oncoprotein. Clin Cancer Res. 2010;16(19):4711–4721. | ||
Rantala H, Mannonen L, Ahtiluoto S, et al. Human herpesvirus-6 associated encephalitis with subsequent infantile spasms and cerebellar astrocytoma. Dev Med Child Neurol. 2000;42(6):418–421. | ||
Shao Q, Lin Z, Wu X, et al. Transcriptome sequencing of neurologic diseases associated genes in HHV-6A infected human astrocyte. Oncotarget. 2016;7(30):48070–48080. | ||
Kashanchi F, Araujo J, Doniger J, et al. Human herpesvirus 6 (HHV-6) ORF-1 transactivating gene exhibits malignant transforming activity and its protein binds to p53. Oncogene. 1997;14(3):359–367. | ||
Doniger J, Muralidhar S, Rosenthal LJ. Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate. Clin Microbiol Rev. 1999;12(3):367–382. | ||
Schleimann MH, Møller JM, Kofod-Olsen E, Höllsberg P. Direct repeat 6 from human herpesvirus-6B encodes a nuclear protein that forms a complex with the viral DNA processivity factor p41. PLoS One. 2009;4(10):e7457. | ||
Borenstein R, Zeigerman H, Frenkel N. The DR1 and DR6 first exons of human herpesvirus 6A are not required for virus replication in culture and are deleted in virus stocks that replicate well in T-cell lines. J Virol. 2010;84(6):2648–2656. | ||
Chi J, Gu B, Zhang C, et al. Human herpesvirus 6 latent infection in patients with glioma. J Infect Dis. 2012;206(9):1394–1398. | ||
Li L, Gu B, Zhou F, et al. Human herpesvirus 6A infects human embryonic fibroblasts and induces G2/M arrest and cell death. J Med Virol. 2012;84(4):657–663. | ||
Wang S, Ma G, Zhu H, et al. miR-107 regulates tumor progression by targeting Nf1 in gastric cancer. Sci Rep. 2016;6:36531. | ||
Albini A, Tosetti F, Li VW, Noonan DM, Li WW. Cancer prevention by targeting angiogenesis. Nat Rev Clin Oncol. 2012;9(9):498–509. | ||
Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473(7347):298–307. | ||
Kashanchi F, Thompson J, Sadaie MR, et al. Transcriptional activation of minimal HIV-1 promoter by ORF-1 protein expressed from the SalI-L fragment of human herpesvirus 6. Virology. 1994;201(1):95–106. | ||
Caruso A, Caselli E, Fiorentini S, et al. U94 of human herpesvirus 6 inhibits in vitro angiogenesis and lymphangiogenesis. Proc Natl Acad Sci U S A. 2009;106(48):20446–20451. | ||
Ala-Aho R, Kähäri VM. Collagenases in cancer. Biochimie. 2005;87(3–4):273–286. | ||
Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. | ||
Jackson HW, Defamie V, Waterhouse P, Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat Rev Cancer. 2017;17(1):38–53. | ||
Akiyama SK, Olden K, Yamada KM. Fibronectin and integrins in invasion and metastasis. Cancer Metastasis Rev. 1995;14(3):173–189. | ||
Totzke G, Essmann F, Pohlmann S, Lindenblatt C, Jänicke RU, Schulze-Osthoff K. A novel member of the IkappaB family, human IkappaB-zeta, inhibits transactivation of p65 and its DNA binding. J Biol Chem. 2006;281(18):12645–12654. | ||
Harada M, Qin Y, Takano H, et al. G-CSF prevents cardiac remodeling after myocardial infarction by activating the JAK–STAT pathway in cardiomyocytes. Nat Med. 2005;11(3):305–311. | ||
Kodama H, Fukuda K, Pan J, et al. Leukemia inhibitory factor, a potent cardiac hypertrophic cytokine, activates the JAK/STAT pathway in rat cardiomyocytes. Circ Res. 1997;81(5):656–663. | ||
Čokić VP, Mitrović-Ajtić O, Beleslin-Čokić BB, et al. Proinflammatory cytokine IL-6 and JAK–STAT signaling pathway in myeloproliferative neoplasms. Mediators Inflamm. 2015;2015:1–13. | ||
David M, Ford D, Bertoglio J, Maizel AL, Pierre J. Induction of the IL-13 receptor alpha2-chain by IL-4 and IL-13 in human keratinocytes: involvement of STAT6, ERK and p38 MAPK pathways. Oncogene. 2001;20(46):6660–6668. | ||
Zanotto-Filho A, Gonçalves RM, Klafke K, et al. Inflammatory landscape of human brain tumors reveals an NFκB dependent cytokine pathway associated with mesenchymal glioblastoma. Cancer Lett. 2017;390:176–187. | ||
Stechishin OD, Luchman HA, Ruan Y, et al. On-target JAK2/STAT3 inhibition slows disease progression in orthotopic xenografts of human glioblastoma brain tumor stem cells. Neuro Oncol. 2013;15(2):198–207. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.