Back to Journals » OncoTargets and Therapy » Volume 14
Downregulation of TAF9B by miR-7-5p Inhibits the Progression of Osteosarcoma
Authors Gu W, Chen P, Ren P, Wang Y, Li X, Gong M
Received 28 May 2020
Accepted for publication 2 February 2021
Published 30 April 2021 Volume 2021:14 Pages 2917—2927
DOI https://doi.org/10.2147/OTT.S264786
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Wanli Gu,1 Peng Chen,1 Peng Ren,1 Yanhai Wang,2 Xiaobing Li,3 Mingzhi Gong1
1Department of Orthopaedics, The Second Hospital of Shandong University, Jinan, Shandong, People’s Republic of China; 2Obstetrical Department, The Second Hospital of Shandong University, Jinan, Shandong, People’s Republic of China; 3Institute of Basic Medicine, The First Affiliated Hospital of Shandong First Medical University, Jinan, Shandong Province, People’s Republic of China
Correspondence: Mingzhi Gong
Department of Orthopaedics, The Second Hospital of Shandong University, 247 Beiyuan Street, Jinan, 250033, Shandong, People’s Republic of China
Tel +86 17660081696
Email [email protected]
Background: Osteosarcoma (OS) is a malignant bone tumor with high metastatic potential. As a regulatory factor of apoptosis, TATA-box binding protein (TBP) associated factor 9B (TAF9B) is rarely studied in tumors.
Methods: We investigated the role and mechanism of TAF9B in OS cells by overexpression and knockdown. CCK8, colony formation, transwell, and flow cytometry analysis were performed to detect proliferation, migration, invasion, and apoptosis.
Results: TAF9B overexpression promotes the proliferation, migration, and invasion of OS cells, while TAF9B knockdown gives the opposite result. TAF9B inhibits apoptosis by upregulating Bcl-2 and downregulating Bax and Cleaved-caspase-3. Through starBase analysis, it was found that miR-7-5p can bind to the 3ʹUTR region of TAF9B, which is further confirmed by the dual luciferase reporter system assay. MiR-7-5p downregulates the expression of TAF9B in MG63 and U2OS cells. The proliferation and invasion of OS cells are inhibited after miR-7-5p mimics transfection and are promoted after miR-7-5p inhibitor transfection. TAF9B rescues the inhibitory effect of miR-7-5p on OS cells. TAF9B also activates the AKT/mTOR signaling pathway.
Conclusion: According to our results, miR-7-5p inhibits the translation of TAF9B and then suppresses growth and metastasis through the AKT/mTOR signaling pathway in OS cells, thereby indicating the potential value of miR-7-5p and TAF9B as therapeutic targets for human OS.
Keywords: TAF9B, osteosarcoma, miR-7-5p, 3ʹUTR
Introduction
Osteosarcoma (OS) is a malignant bone tumor with high metastatic potential. Over the past three decades, research aimed at improving the effectiveness of OS chemotherapy has progressed slowly.1 One of the important reasons for the lack of new treatment strategies is the extremely complex genetic background of OS. Up until now, no specific driving genes and related pathogenesis have been identified in OS. Therefore, it is very important to investigate the pathogenesis and progression of OS.
TATA-box binding protein (TBP) associated factor 9B (TAF9B) is a subunit of transcription factor IID (TFIID), which assists RNA polymerase II to initiate transcription.2 At present, there are few reports about the function of TAF9B in cells and diseases. Current research shows that TAF9B participates in the regulation of cell apoptosis and autophagy through the p53 signaling pathway. Frontini et al reported for the first time that overexpressed TAF9B can protect p53 from degradation.2 MiR-146a can target TAF9B and indirectly induce the degradation of P53 to inhibit apoptosis and promote autophagy in cardiomyocytes.3 However, TAF9B is rarely studied in tumors.
In recent years, the research on the role of miR-7-5p in tumors’ biological behavior has gradually increased. MiR-7-5p operates as a tumor suppressor in multiple malignant tumors, including colorectal cancer, breast cancer, lung cancer, head and neck cancer, pancreatic cancer, gastric cancer, bladder cancer, esophageal squamous cell carcinoma, and ovarian cancer.4–11 In 2018, miR-7-5p was proven to inhibit tumor growth of osteosarcoma, but its downstream mechanism remains unclear.12 In the present research, we found that TAF9B was a downstream target of miR-7-5p and we explored the specific mechanism by which TAF9B/miR-7-5p promoted OS progression in cell lines cultured in vitro.
Materials and Methods
Cell Lines and Transfection
Human Osteoblast cell line hFOB 1.19 and OS cell lines MG-63 and U2OS were all purchased from the Cell Resource Center, Shanghai Institutes for Biological Sciences, the Chinese Academy of Sciences. DMEM with 10% FBS was used for the cell culture.
After about 80% confluence of cells, transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. TAF9B specific siRNA, siRNA negative control, miR-7-5p mimics, and miR-7-5p inhibitor were all purchased from RiboBio Co. Ltd. (Guangzhou, China). The TAF9B overexpression plasmid was purchased from YouBio Technology Co. Ltd. (Hunan, China). The sequence of TAF9B-specific siRNA was 5ʹ- GUUCAAAAUGUUCUGAUUAAU −3ʹ; the sequence of siRNA negative control was 5ʹ-GGUCUCCCAAACGGUCUAUTT-3ʹ.
Real-Time Fluorescence Quantitative PCR Analysis (qPCR)
After transfection for 24 h, total RNA was extracted from cells with an Ultrapure RNA Kit (Beijing CoWin Biotech Co. Ltd., Beijing, China). Reverse transcription PCR and qPCR were performed successively using HiFiScript cDNA Synthesis Kit and UltraSYBR Mixture, respectively (Beijing CoWin Biotech Co. Ltd., Beijing, China). The expression level of TAF9B was analyzed using the 2−ΔΔct method. All qPCR analysis was performed in triplicate. The primers, TAF9B-F5ʹ-GCAGATTCCACCTTCTCAGTCC-3ʹ, TAF9B-R5ʹ-CTGTGACGAAACCATGTTGGTGG-3ʹ, β-actin-F5ʹ-CCCGAGCCGTGTTTCCT-3ʹ, and β-actin-R5ʹ-GTCCCAGTTGGTGACGATGC-3ʹ were used in the present research.
Western Blotting
Protein was extracted from MG-63 and U2OS cells after transfection for 48 h. SDS-PAGE was performed to separate the protein sample (20 μg protein/lane). The samples were transferred to a polyvinylidene fluoride (PVDF) membrane, and then incubated with 5% non-fat milk, primary antibodies, and secondary antibodies successively. An ECL kit was used for visualization. The relative expression of the target protein was analyzed using Image J software (National Institutes of Health Bethesda, USA) with GAPDH as a loading control. Primary antibodies anti-TAF9B (1:1000, cat#ab133239, Abcam, UK), anti-Bcl-2 (1:2000, cat#26593-1-AP, ProteinTech Group, USA), anti-Bax (1:2000, cat#50599-2-Ig, ProteinTech Group, USA), anti-Cleaved-caspase-3 (1:1000, cat# 19677-1-AP, ProteinTech Group, USA), anti-AKT (1:3000, cat# 10176-2-AP, ProteinTech Group, USA), anti-p-AKT (1:3000, cat#66444-1-Ig, ProteinTech Group, USA), anti-mTOR (1:2000, cat#ab2732, Abcam, UK), anti-p-mTOR (1:1000, cat#ab109268, Abcam, UK), and anti-GAPDH (1:5000, cat#10494-1-AP, ProteinTech Group, USA) were used in this research.
Cell Proliferation Assay
The proliferation of MG-63 and U2OS cells was determined by using a Cell Counting Kit-8 (CCK-8) (Beijing Solarbio Science and Technology Co., Ltd., China). After the transfection with siRNA-TAF9B, siRNA negative control, miR-7-5p mimics, or miR-7-5p inhibitor, cells were transferred into a 96-well plate (1000 cells/well). Cell proliferation was detected after incubation for 0, 24, 48, and 72 h. CCK-8 regent (10 μL/well) was added to cells and incubated for 1.5 h at 37 °C. The optical density (OD) value was measured at 450 nm using a microplate reader.
Colony formation assay was performed to further determine the proliferation of MG-63 and U2OS cells. After transfection, 200 cells were seeded into a 3.5 cm plate and incubated for 14 days at 37 °C. Then, cells were fixed with 4% paraformaldehyde for 20 min and subsequently stained with crystal violet for 30 min. Colony number was counted in 5 random fields/plate.
Transwell Assay
100 μL Matrigel dissolved overnight was added to the upper chamber of the transwell and incubated at 37 °C for 6 h. After 24 h of transfection, 100 μL cell suspension (serum-free medium) (1×105 cells) was added to the upper chamber, and 500 uL complete medium was added to the lower chamber. After overnight culture at 37 °C, the invasive cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 20 min. 5 fields were randomly photographed under the microscope.
Migration transwell was similar to that of invasion assay. The transwell chamber had no Matrigel, and the cell number was 5000/well.
Dual Luciferase Reporter System Assay
The 3ʹUTR and its mutation sequence of TAF9B were cloned into pGL3.0 vector. The transfection was performed when 293T cells reached 80% confluence. The cells were divided into four groups: pGL3.0-TAF9B-wt, pGL3.0-TAF9B-wt + miR-7-5p, pGL3.0-TAF9B-mut, pGL3.0-TAF9B-mut + miR-7-5p. After 48 h of culture, a double luciferase system (Promega, USA) was used to detect the fluorescence value of each group.
Flow Cytometry Analysis
The apoptosis of MG-63 and U2OS cells was analyzed using Flow Cytometry with an Annexin V-FITC Apoptosis Detection Kit (Beijing Solarbio Science and Technology Co., Ltd., China). After 24 h of transfection, cells were stained and incubated following the manufacturer’s instructions. FlowJo software was used for data analysis.
Statistical Analyses
GraphPad Prism 8 was used in the present research for the statistical analysis. Student’s t-test was used for the comparison between two groups, and one-way ANOVA was used for the comparison between multiple groups. All experiments were performed in triplicate. p<0.05 represents statistical significance.
Results
TAF9B Promotes the Proliferation in OS Cells
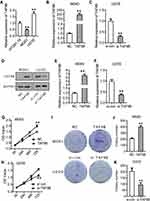
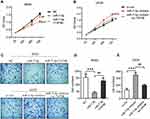
First, we detected the expression of TAF9B using qPCR in OS cell lines MG-63 and U2OS with osteoblast cell line hFOB 1.19 as a control. The results showed that TAF9B was lowly expressed in MG-63 and highly expressed in U2OS (Figure 1A). Thus, we transfected the TAF9B overexpression plasmid into MG-63 cells with the empty plasmid pcDNA3.1 as a control. Moreover, we transfected si-TAF9B into U2OS cells with the nonspecific siRNA as a control to investigate the role of TAF9B in OS cells. Form the results of qPCR, TAF9B expression was upregulated after the transfection of TAF9B overexpression plasmid in MG-63 cells (Figure 1B), and TAF9B expression was downregulated after the transfection of si-TAF9B in U2OS cells (Figure 1C). From the results of the Western blot, the protein level of TAF9B increased after the transfection of TAF9B overexpression plasmid in MG63 cells (Figure 1D and E), while the TAF9B protein level declined after the transfection of si-TAF9B in U2OS cells (Figure 1D and F).
We detected the proliferation of MG-63 and U2OS cells using CCK-8 and colony formation assays. As shown in Figure 1G and H, the OD450 value increased after the overexpression of TAF9B compared with the NC group in MG-63 cells, while the OD450 value decreased after the knockdown of TAF9B compared with the si-con group in U2OS C cells. In colony formation assay, the colony number increased markedly in the TAF9B group compared with the NC group in MG-63 cells and it declined in the si-TAF9B group compared with the si-con group in U2OS cells, which was consistent with the results of CCK-8 assay (Figure 1I–K). These data proved that TAF9B promoted the proliferation of OS cells in vitro.
TAF9B Promotes the Migration and Invasion in OS Cells
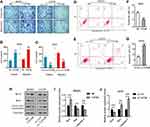
The transwell assay was performed to determine the migration and invasion of MG-63 and U2OS cells. As shown in Figure 2A–C, the migrated cell number increased after the overexpression of TAF9B (TAF9B group, 530±42; NC group, 245±21) in MG-63 cells and decreased after the knockdown of TAF9B compared with the control group (si-TAF9B group, 90±14; si-con group, 215±35) in U2OS cells. Consistent with the results of the migration assay, the invasive cell number increased in the TAF9B overexpression cells (TAF9B group, 435±35; NC group, 200±28) in MG-63, while the invasive cell number decreased in the TAF9B knockdown cells (si-TAF9B group, 43±6; si-con group, 153±20) in U2OS. Our results indicated that TAF9B could promote the migration and invasion in OS cells.
TAF9B Inhibits the Apoptosis in OS Cells
Next, flow cytometry analysis was performed to determine the apoptosis in MG-63 and U2OS cells. As shown in Figure 2D–G, the percentage of apoptotic cells declined in the TAF9B overexpression group compared with the NC group in MG-63, while the percentage of apoptotic cells increased in the TAF9B knockdown group compared with the si-con group in cells U2OS cells (Figure 2).
We then measured the expression levels of cell apoptosis relative protein using Western blot assay. As shown in Figure 2H–J, Bax and Cleaved-caspase-3 were downregulated by TAF9B overexpression in MG-63 cells and upregulated by TAF9B knockdown in U2OS cells. Bcl-2 was upregulated by TAF9B overexpression in MG-63 cells and downregulated by TAF9B knockdown in U2OS cells. The above results proved that TAF9B inhibited cell apoptosis in human OS.
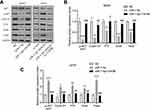
TAF9B Activates the AKT/mTOR Signaling Pathway in OS Cells
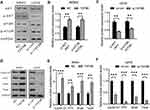
To explore the mechanism by which TAF9B promotes the growth of OS cells, we detected the AKT/mTOR signaling pathway using Western blot assay, which was the critical pathway regulating cell proliferation and apoptosis. As shown in Figure 3A–C, the phosphorylation of AKT and mTOR increased after the overexpression of TAF9B in MG-63 cells and decreased after the knockdown of TAF9B in U2OS cells. Then, we detected the expression of key proteins associated with proliferation and metastasis. As shown in Figure 3D–F, the levels of Cyclin D1, p70, Snail, and Twist were up-regulated after the overexpression of TAF9B in MG-63 cells and decreased after the knockdown of TAF9B in U2OS cells.
MiR-7-5p Inhibits the Expression of TAF9B Through Targeting Its 3ʹUTR Region in OS Cells
We further analyzed the binding sites on TAF9B 3ʹUTR region with miRNAs using the starBase online tool (http://starbase.sysu.edu.cn/starbase2/index.php).13 The results showed that there was a binding site between TAF9B 3ʹUTR and miR-7-5p in the analysis results of three databases (TargetScan, RNA22, and miRanda sites).14–16
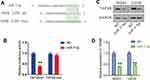
In order to verify their binding in cells, the wild-type (UCUUCCA) and mutant sequences (AGAAGGU) of the binding site in the TAF9B 3ʹUTR region were inserted into pGL3.0 plasmid (Figure 4A) to generate TAF9B-3ʹUTR (wt) and TAF9B-3ʹUTR (mut) luciferase reporter plasmids, respectively. Then, dual luciferase reporter system assay was performed. As shown in Figure 4B, relative luciferase activity declined significantly after the transfection of miR-7-5p mimics and TAF9B-3ʹUTR (wt) compared with the transfection of TAF9B-3ʹUTR (wt), while co-transfection of TAF9B-3ʹUTR (mut) and miR-7-5p did not affect the relative luciferase activity. As such, we concluded that miR-7-5p bound to the TAF9B-3ʹUTR and then weakened the fluorescence in cells, thereby indicating that TAF9B was a direct target of miR-7-5p.
We further detected the effect of miR-7-5p on TAF9B expression in the cells transfected with miR-7-5p mimics using Western blot assay. As shown in Figure 4C and D, TAF9B expression was significantly inhibited by miR-7-5p mimics both in MG-63 and U2OS cells.
MiR-7-5p Inhibits the Proliferation and Invasion in OS Cells
We subsequently investigated the effect of miR-7-5p through the transfection of miR-7-5p mimics in MG-63 cells and inhibitor in U2OS cells. From the results of CCK-8, and following the transfection for 72 h, the OD450 declined significantly in the miR-7-5p group compared with the NC group in MG-63 cells and increased in the miR-7-5p inhibitor group compared with the si-con group in U2OS cells (Figure 5A and B). The invasive cell number was also inhibited by miR-7-5p mimics in MG-63 cells and promoted by miR-7-5p inhibitor in U2OS cells (Figure 5C–E). These data indicate that miR-7-5p could inhibit the proliferation and invasion in human OS cells.
TAF9B Rescues the Inhibition of Proliferation and Invasion from miR-7-5p in OS Cells
Finally, we used miR-7-5p+TAF9B cells generated by the transfection of miR-7-5p mimics+TAF9B overexpression plasmid and miR-7-5p-inhibitor+si-TAF9B cells generated by the transfection of miR-7-5p-inhibitor and siRNA-TAF9B to detect the effect of TAF9B on miR-7-5p. As shown in Figure 5, the inhibition of proliferation and invasion induced by miR-7-5p in MG-63 cells was blocked by the overexpression of TAF9B, while the enhancement of proliferation and invasion induced by miR-7-5p inhibitor was rescued by TAF9B knockdown in U2OS cells.
Then, we detected the expression levels of AKT, Cyclin D1, p70, Snail, and Twist. As shown in Figure 6, the inhibition of AKT, Cyclin D1, p70, Snail, and Twist expression induced by miR-7-5p in MG-63 cells was blocked by the overexpression of TAF9B, while the enhancement of these markers induced by miR-7-5p inhibitor was rescued by TAF9B knockdown in U2OS cells. Our results proved that miR-7-5p inhibited proliferation and invasion through targeting TAF9B in human OS cells.
Discussion
In the present research, we focused on investigating the role and mechanism of TAF9B as a cancer-promoting factor in human OS cells. In 2005, TAF9B was first identified as a subunit of TFIID, and knockdown of TAF9B led to apoptotic cell death in human HeLa cells.2 Since then, several reports have shown that TAF9B regulates cell survival through the p53 signaling pathway. In breast cancer patients, the loss of heterozygosity (LOH) in TAF9B is correlated to prognosis, suggesting the potential value of TAF9B as a prognostic marker.17 TAF9B forms a regulatory loop with SNHG1 and sno-miR-28, thus regulating the stability of p53 as well as the downstream pathway.18 Moreover, TAF9B is also involved in the transcriptional regulation during embryonic germ cell development.19 Herrera et al found that TAF9B is preferentially related to PCAF in neurons rather than the canonical TFIID complex, indicating that TAF9B may be involved in other transcriptional regulation pathways.20
In our study, we first detected that effect of TAF9B on the biological behavior of OS cells. TAF9B could promote proliferation, migration, and invasion and likewise inhibit apoptosis in MG-63 and U2OS cells. TAF9B also promoted the phosphorylation levels of AKT and mTOR, suggesting that TAF9B targeted and activated the AKT/mTOR signaling pathway in OS cells.
AKT is a crucial molecule of the cell survival pathway, the PI3K/AKT signaling pathway, and has been defined as an oncogene.21 Its sustained activation is tightly bound to the occurrence and development of tumors.22 AKT promotes the growth, metastasis, and angiogenesis of tumors; it also inhibits cell apoptosis and resists chemotherapy as well as radiotherapy by phosphorylating mTOR, GSK3, the FoxO family, and other substrates.23–25 We found that TAF9B also regulated downstream target proteins of AKT, including Bcl-2, Bax, and Caspase-3 to regulate tumor cell apoptosis. Therefore, our results proved that TAF9B could promote cell proliferation and survival by activating the AKT pathway in human OS cells. In addition, AKT is involved in the regulation of the cell cycle, apoptosis, and DNA repair through p53, which is the downstream target of TAF9B in several reports.26,27 Thus, we hypothesize that TAF9B may play a role in the cell cycle and DNA repair through the AKT/p53 signaling pathway in OS cells, which needs to be further verified.
MiRNAs widely involve in tumor progression by regulating mRNA at the post-transcriptional level.28 Mature single-stranded miRNAs combine to RNA-induced silencing complex (RISE) in a complete or incomplete pairing manner and suppress the initiation and extension of mRNA translation, which ultimately inhibits gene expression.29 In the present research, and according to the results of starBase analysis and dual luciferase reporter assay, miR-7-5p combined with the 3 ‘UTR region of TAF9B. MiR-7-5p could significantly inhibit the expression of TAF9B protein and then inhibit the proliferation and invasion in OS cells.
MiR-7-5p is a non-coding small molecule RNA first reported by Lagos, located on human chromosome 9q21.30 Mature miR-7-5p is composed of 23 nucleotides and plays an important regulatory role in different signaling pathways through multiple target genes, such as epidermal growth factor receptor (EGFR).31,32 Multiple studies have confirmed that miR-7-5p is downregulated in various tumors and involved in the inhibition of cell proliferation and metastasis by regulating target molecules.32 Our results also showed that miR-7-5p operated as a tumor suppressor in OS. In addition, miR-7-5p rescues the resistance to BRAFi via the inhibition of the PI3K/AKT signaling pathways in human melanoma.8 In a diabetic rat model, MiR-7 inhibits the proliferation of retinal capillary endothelial cells through regulating the PI3K/AKT/VEGF pathway.33 We proved that TAF9B both targeted and activated the AKT/mTOR signaling pathway in OS cells. Combined with previous studies, we hypothesize that miR-7-5p could target the AKT/mTOR signaling pathway through TAF9B in OS cells, which needs further investigation.
In conclusion, we have proven that TAF9B promotes proliferation, survival, and metastasis in OS cells through activating the AKT/mTOR signaling pathway. MiR-7-5p inhibits cell growth by binding and targeting TAF9B in OS. These results enrich the regulatory network of OS development and provide new potential targets for targeted therapy of human OS.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Statement
Ethics approval and consent to participate This research study was approved by the Second Hospital of Shandong University.
Consent for Publication
All the authors agree to the publication clause.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests exist.
References
1. Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65.
2. Frontini M, Soutoglou E, Argentini M, et al. TAF9b (Formerly TAF9L) Is a Bona Fide TAF that has unique and overlapping roles with TAF9. Mol Cell Biol. 2005;25(11):4638–4649. doi:10.1128/MCB.25.11.4638-4649.2005
3. Pan JA, Tang Y, Yu JY, et al. miR-146a attenuates apoptosis and modulates autophagy by targeting TAF9b/P53 pathway in doxorubicin-induced cardiotoxicity. Cell Death Dis. 2019;10(9):668. doi:10.1038/s41419-019-1901-x
4. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-0454-8
5. Yang W, Yang X, Wang X. Silencing CDR1as enhances the sensitivity of breast cancer cells to drug resistance by acting as a miR-7 sponge to down-regulate REGγ. J Cell Mol Med. 2019;23(8):4921. doi:10.1111/jcmm.14305
6. Kalinowski FC, Giles KM, Candy PA, et al. Regulation of epidermal growth factor receptor signaling and erlotinib sensitivity in head and neck cancer cells by miR-7. PLoS One. 2012;7(10):e47067. doi:10.1371/journal.pone.0047067
7. Li YJ, Wang CH, Zhou Y, et al. TLR9 signaling repressed tumor suppressor miR-7 expression through up-regulation of HuR in human lung cancer cells. Cancer cell int. 2013;13(1):90.
8. Pan H, Li T, Jiang Y, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on Gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119(1):440–446.
9. Xu G, Yang H, Liu M, et al. lncRNA TINCR facilities bladder cancer progression via regulating miR‑7 and mTOR. Mol Med Rep. 2020;22(5):4243–4253.
10. Meng X, Joosse SA, Müller V, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. 2015;113(9):1358–1366.
11. Li R, Ke S, Meng FK, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9(8):1–3.
12. Xu B, Yang T, Wang Z. CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. 2018;10:4871.
13. Li JH, Liu S, Zhou H, et al. Starbase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale clip-seq data. Nucleic Acids Res. 2013;42(D1):D92.
14. Vikram A, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;12:e05005.
15. Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the Identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1217.
16. Betel D, Koppal A, Agius P, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi:10.1186/gb-2010-11-8-r90
17. Deryusheva IV, Tsygano MM, Garbukov EY, et al. Genome-wide association study of loss of heterozygosity and metastasis-free survival in breast cancer patients. Exp Oncol. 2017;39(2):145–150. doi:10.31768/2312-8852.2017.39(2):145-150
18. Yu F, Bracken CP, Pillman KA, et al. p53 represses the oncogenic Sno-MiR-28 derived from a SnoRNA. PLoS One. 2015;10(6):e0129190. doi:10.1371/journal.pone.0129190
19. Gura MA, Mikedis MM. Dynamic and regulated TAF gene expression during mouse embryonic germ cell development. PLoS gene. 2020;16(1):e1008515.
20. Herrera FJ, Yamaguchi T, Roelink H, et al. Core promoter factor TAF9B regulates neuronal gene expression. Elife. 2014;3:e02559. doi:10.7554/eLife.02559
21. Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase|[ndash]|AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi:10.1038/nrc839
22. Zhang J, Yu X-H, Yan Y-G, et al. PI3K/Akt signaling in osteosarcoma. Clin Chimica Acta. 2015;444:182–192. doi:10.1016/j.cca.2014.12.041
23. Memmott RM, Dennis PA. Akt-dependent and -independent mechanisms of mTOR regulation in cancer. Cell Signal. 2009;21(5):656–664. doi:10.1016/j.cellsig.2009.01.004
24. Hua K, Li Y, Zhao Q, et al. Downregulation of Annexin A11 (ANXA11) inhibits cell proliferation, invasion, and migration via the AKT/GSK-3β Pathway in Gastric Cancer. Med Sci Monitor Int Med J Exp Clin Res. 2018;24:149–160.
25. Lu H, Zhang LH, Yang L, et al. The PI3K/Akt/FOXO3a pathway regulates regeneration following spinal cord injury in adult rats through TNF-α and p27kip1 expression. Int J Mol Med. 2018;41(5):2832.
26. Chen L, Tang J, Feng Y, et al. ADAMTS9 is silenced by epigenetic disruption in colorectal cancer and inhibits cell growth and metastasis by regulating Akt/p53 Signaling. Cell Physiol Biochem. 2017;44(4):1370–1380.
27. Kim EJ, Kim GT, Kim BM, et al. Apoptosis-induced effects of extract from Artemisia annua Linné by modulating PTEN/p53/PDK1/Akt/signal pathways through PTEN/p53-independent manner in HCT116 colon cancer cells. BMC Complement Altern Med. 2017;17(1). doi:10.1186/s12906-017-1702-7
28. Salehi M, Sharifi M. Exosomal miRNAs as novel cancer biomarkers: challenges and opportunities. J Cell Physiol. 2018;233(9):6370–6380. doi:10.1002/jcp.26481
29. Ferreira HJ, Esteller M. Non-coding RNAs, epigenetics, and cancer: tying it all together. Cancer Metastasis Rev. 2018;37(1):55–73.
30. Lagos-Quintana M. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi:10.1126/science.1064921
31. Eslamizadeh S, Heidari M, Agah S, et al. The role of MicroRNA signature as diagnostic biomarkers in different clinical stages of colorectal cancer. Cell J. 2018;20(2):220–230. doi:10.22074/cellj.2018.5366
32. Jessica H, Kalinowski FC, Epis MR, et al. Clinical Potential of microRNA-7 in Cancer. J Clin Med. 2015;4(9):1668–1687. doi:10.3390/jcm4091668
33. Cao YL, Liu DJ, Zhang HG. MiR-7 regulates the PI3K/AKT/VEGF pathway of retinal capillary endothelial cell and retinal pericytes in diabetic rat model through IRS-1 and inhibits cell proliferation. Eur Rev Med Pharmacol Sci. 2018;22(14):4427–4430.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.