Back to Journals » OncoTargets and Therapy » Volume 11
Downregulation of STARD8 in gastric cancer and its involvement in gastric cancer progression
Authors Zhang S, Chang X , Ma J, Chen J, Zhi Y, Li Z, Dai D
Received 19 October 2017
Accepted for publication 7 March 2018
Published 21 May 2018 Volume 2018:11 Pages 2955—2961
DOI https://doi.org/10.2147/OTT.S154524
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Shuanglong Zhang,1,2,* Xiaojing Chang,1–3,* Jinguo Ma,2,4 Jing Chen,1 Yu Zhi,2 Zhenhua Li,2 Dongqiu Dai1,2
1Department of Gastrointestinal Surgery, The Fourth Affiliated Hospital, China Medical University, Shenyang, People’s Republic of China; 2Cancer Center, The Fourth Affiliated Hospital, China Medical University, Shenyang, People’s Republic of China; 3Department of Radiotherapy, The Second Hospital of Hebei Medical University, Shijiazhuang, People’s Republic of China; 4Hulun Buir People’s Hospital, Hulun Buir Medical School in Nationalities University of Inner Mongolia, Hulun Buir, People’s Republic of China
*These authors contributed equally to this work
Objective: Rho-GTPases play a pivotal role in a wide variety of signal transduction pathways and are associated with a great number of human carcinomas. STARD8, which is a Rho-GTPase-activating protein, has been proposed as a tumor suppressor gene, but its role in gastric cancer remains elusive. In this study, we investigate the expression of STARD8 in gastric cancer and its association with gastric cancer progression.
Materials and methods: One normal gastric mucosa cell line for example GES1 and six human gastric cancer cell lines such as AGS, MGC803, MKN45, SGC7901, HGC27 and BGC823 were utilized to analyze STARD8 mRNA and protein levels by reverse transcription polymerase chain reaction (RT-PCR) and Western blot. A total of 70 paired gastric tissues including corresponding nonmalignant gastric tissues and cancer tissues were utilized to analyze the protein expression of STARD8 using immunohistochemistry, and the correlation between STARD8 level and clinicopathological features was also evaluated.
Results: STARD8 was found to be downregulated in primary gastric cancer cells and tissues compared with the normal gastric mucosa cell line, GES1, and corresponding nonmalignant gastric tissues, while its decreased expression was significantly associated with TNM stage, lymph node metastasis and differentiation (p<0.05).
Conclusion: There is significantly decreased expression of STARD8 in gastric cancer cells and tissues, and its expression may contribute to gastric tumorigenesis.
Keywords: gastric cancer, STARD8, Rho-GTPase-activating proteins, metastasis
Introduction
Gastric cancer (GC) has been predicted to be the leading cause of all deaths worldwide, especially in China, and it is the second most common cause of cancer-related death.1 Although curable if detected early, most patients are diagnosed with advanced GC. Current therapeutic strategies, including surgery and combination chemotherapies, are shown to provide only modest survival benefits for patients with advanced GC. Thus, despite a steady decline in global prevalence, GC remains a disease of outstanding mortality.2
Rho-GTPases are important molecular switches that control a wide variety of signal transduction pathways in all eukaryotic cells.3 They play a pivotal role in regulating the actin cytoskeleton and also influence cell polarity, microtubule dynamics, membrane transport pathways and transcription factor activities.4,5 The activity of Rho-GTPases varies between the active GTP-bound state and the inactive GDP-bound state and regulates a number of cellular processes such as cytoskeleton reorganization, gene transcription and cell cycle progression.3 Upregulation of the expression and activity of Rho-GTPases have been confirmed to be associated with a great number of human carcinomas.6,7 Rho-GTPase-activating proteins (Rho-GAPs) have three subgroups: deleted in liver cancer gene 1-3 (DLC1-3).8 DLC-1 and DLC-2 genes have been shown to be downregulated in cancer cells and work to inhibit cancer cell proliferation.9–12 DLC-3, also known as STARD8 and KIAA0189, is another Rho-GAP that maps to chromosome Xq13, which was first isolated from a human myeloid cell line library in 1996. STARD8 consists of a sterile alpha motif (SAM), GAP and START (steroidogenic acute regulatory protein [StAR]-related lipid transfer) domain which has been reported to be downregulated in some carcinomas, such as breast, ovarian, liver and prostate cancer cell lines.13,14 However, the role of STARD8 in gastric carcinoma is still unclear.
In this study, we investigated the profiles of STARD8 in several human GC cell lines and tissues. We collected data regarding mRNA and protein levels of STARD8 from a cohort of patients with GC (n=70) to analyze the possible role of STARD8 in patients with GC.
Materials and methods
Human cell lines
The human GC cell lines AGS, MGC803, MKN45, SGC7901, HGC27 and BGC823 were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China); one immortalized normal gastric cell line, GES1, that was approved by the institutional review board of China Medical University was obtained from the Oncology Institute of China Medical University and maintained as recommended. Cells were cultured in RPMI-1640 (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (Thermo Fisher Scientific) in a humidified atmosphere of 5% CO2 at 37°C.
Tissue samples
Specimens were collected from 70 patients with gastric carcinoma who underwent gastrectomy at the Cancer Institute of China Medical University, Shenyang, China, between January 2008 and April 2010. This study was approved by the institutional review board of China Medical University, and each patient signed an informed consent form to participate in this study. All GC cases were pathologically confirmed, and the normal tissues were at least 5 cm away from cancer lesions. A total of 24 patients were women and 46 patients were men, and the mean age was 62 years (range: 30–78 years).
RNA extraction and reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from fresh frozen human GC tissues using the TRIzol reagent as described by the manufacturer (Thermo Fisher Scientific). cDNA was synthesized from 2 μg of total RNA by using oligo(dT)16 as the primer with the Expand Reverse Transcriptase Kit (Fermentas, Vilnius, Lithuania). Polymerase chain reaction (PCR) amplification of STARD8 was performed using the DreamTaq™ DNA Polymerase PCR system (Fermentas), and GAPDH was used as a control. Primers used for RT-PCR are summarized in Table 1.
  | Table 1 Primers used for RT-PCR |
Western blotting
Whole cell lysates were prepared from GC cell lines. Samples were loaded onto 10% sodium dodecyl sulfate (SDS) polyacrylamide gels, transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA) and incubated with primary mouse polyclonal antibody against human STARD8 (Abcam, Cambridge, MA, USA) and rabbit monoclonal antibody against β-actin (Sigma-Aldrich Co., St Louis, MO, USA) overnight at 4°C. Membranes were incubated with a horseradish-peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology Inc., Dallas, TX, USA) for 2 h at room temperature, and immunoreactive protein bands were visualized with a chemiluminescent detection kit (enhanced chemiluminescence [ECL]; Thermol Biotech Inc., Rockford, IL, USA). Signal intensities were quantified and analyzed as described in the following sections. Each experiment was repeated three times.
Immunofluorescence staining
The expression of STARD8 in six gastric carcinoma cell lines and one normal gastric mucosa cell line, GES1, was analyzed by immunofluorescence microscopy. In brief, the cells were cultured to confluence on collagen-coated glass coverslips and then fixed in 4% paraformaldehyde (with 50 mM phosphate buffer) in 50% Tris wash buffer (TWB). The glass coverslips were rinsed three times and permeabilized with 1.2% Triton X-100 for 10 min, rinsed three times and incubated with 5% bovine serum albumin (BSA) in 100% TWB for 2 h and then stained for the expression markers using the following primary antibodies: mouse polyclonal antibody against human STARD8 (Abcam) at 1:100 dilution and the rabbit anti-mouse IgG secondary antibodies conjugated with Tritic (red; Zymax, San Francisco, CA, USA). Cells were co-stained with Hoechst 33358 (blue Molecular Devices LLC, Sunnyvale, CA, USA)to visualize nuclei, mounted with mounting medium and viewed by immunofluorescence microscopy.
Immunohistochemistry (IHC)
Slides (4 μm) from all the formalin-fixed paraffin-embedded specimens were made and then dewaxed, rehydrated and incubated in methanol containing 0.3% H2O2 for 10 min to inhibit endogenous peroxidase activity before being blocked with 10% normal goat serum (Maxin, Fuzhou, China) for 30 min and incubating overnight at 4°C with mouse anti-STARD8 (1:1,000; Abcam). Slides were washed in phosphate-buffered saline (PBS), then incubated with biotinylated rabbit anti-mouse IgG for 30 min at room temperature and rinsed with PBS. After being incubated with an avidin–biotin–peroxidase complex for 10 min, sections were stained with 3,3-diaminobenzidine. Normal rabbit serum was used as a negative control.
The result of the IHC for STARD8 was judged based on the extent and intensity of staining as follows: 1) the extent of positive cells was estimated as follows: 0≤5%, 1=6%–25%, 2=26%–50%, 3=51%–75%, 4≥75%; 2) the intensity of staining was judged as follows: 0=achromatic, 1=light yellow, 2=yellow, 3=brown. The score of the extent of positive cells was multiplied by the score of the intensity of staining, and the combined staining score is as follows: (−)=0, (+)=1–4, (++)=5–8, (+++)=9–12.15 Specimens were considered low expression for STARD8 when the score was − or + and were considered high expression when the score was ++ or +++. This semiquantitative analysis was performed by two independent assessors without prior knowledge of the patient outcome.
Statistical analyses
All statistical analyses were performed using SPSS 17.0 software package (SPSS Inc., Chicago, IL, USA). One-way analysis of variance (ANOVA), χ2 test and Student’s t-test were used to generate p-values <0.05 which were considered statistically significant.
Results
Expression of STARD8 was downregulated in human GC cell lines
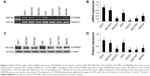
To confirm the role of STARD8 in GC, we first detected the expression of STARD8 in several moderately or poorly differentiated GC cell lines. Results showed that STARD8 mRNA level was substantially downregulated in GC cell lines such as MGC803 (0.79-fold), BGC823 (0.20-fold), AGS (0.60-fold), HGC27 (0.06-fold), SGC7901 (0.21-fold) and MKN45 (0.39-fold) compared with the normal mucosa line GES1 (1-fold as control; Figure 1A and B). Meanwhile, Western blot analysis for these six types of GC cell lines demonstrated that the result of the protein level of STARD8 was similar to the results of STARD8 mRNA level (Figure 1C and D).
Endogenous STARD8 was localized in both nucleus and cytoplasm
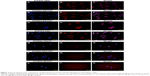
To examine the distribution of endogenous STARD8, we performed immunofluorescence analyses. As indicated, STARD8 localized in both nuclei and cytoplasm, but more strongly localized in nuclei. We compared the differences between the expression levels of STARD8 in six different types of gastric carcinoma cell lines and one normal gastric mucosa cell line (Figure 2). The results revealed that just two of the six cell lines showed significantly reduced expression. This result partly coincides with RT-PCR and Western blot analyses, meaning that the protein expression level of STARD8 was associated with the malignancy of gastric cell lines.
Downregulation of STARD8 in GC tissues and its association with clinicopathological features
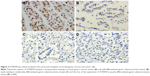
We also examined the expression of STARD8 in GC tissues relative to their pair-matched adjacent non-tumor tissues. Results of IHC confirmed that STARD8 was clearly marked in the “nucleus” and cytoplasm of normal gastric mucosa and cancer cells, predominantly in epithelial cells, while gastric carcinoma samples demonstrated weak or no staining for STARD8 (Figure 3). 28 cases of GC tissues demonstrated no staining which meant that the expression of STARD8 was lost in these 28 cases, whereas all nonneoplastic tissues expressed STARD8, and STARD8 was highly stained in 51 out of 70 (72.9%) nonneoplastic tissues, but only 11 (15.7%) of the GC tissues demonstrated strong staining for STARD8 (p<0.001; Figure 3).
The association of the expression of STARD8 with the clinicopathological parameters of GC patients was summarized in Table 2. The expression of STARD8 had a statistically significant inverse correlation with differentiation (p<0.01), TNM type (p<0.05) and lymph node metastasis (p<0.05). We found that the expression of STARD8 in patients with lymph node metastasis was significantly lower than patients with no lymph node metastasis (p=0.011).
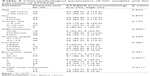  | Table 2 Clinicopathological parameters of GC samples and the expression of STARD8 |
Discussion
Over a decade of intense study about Rho-GAPs has confirmed that Rho-GAP is involved in the regulation of diverse cellular functions and closely correlated with multiple types of carcinomas.16, 17 It has been reported that the three genes deleted in liver cancer genes (DLC1-3) encode Rho-GAPs whose expressions are frequently downregulated or silenced in several human malignancies.13,18 The Rho-GAP DLC-1 (also called STARD12) functions as a tumor suppressor gene in many types of tumors and inhibits the proliferation, migration and tumorigenicity of human carcinoma cells.19–21 DLC-2, a previously identified tumor suppressor gene at 13q12 which encodes the Rho-GAP STARD13, which is homologous to DLC-1, is frequently downregulated in many types of tumors and may suppress cell growth via the regulation of the Raf-1-ERK1/2-p70S6K signaling pathway.22,23 The great similarity among the GAP domains of STARD8 and those of DLC-1 and DLC-2 implies that DLC-3 may also act as a Rho-GAP that is essential for effects on cytoskeletal organization and cell proliferation. To date, studies have shown that STARD8 is downregulated in several different types of cancers, including breast cancer, prostate carcinoma and ovarian neoplasms and contributes to the process of tumorigenesis, whereas its overexpression was shown to inhibit cell proliferation, colony formation and growth.13,14 Mokarram et al24 found that the downregulated expression of STARD8 in colorectal cancer may be due to STARD8 gene promoter methylation. Holeiter et al25 found that DLC-3 depletion could lead to mislocalization of E-cadherin and catenins, which was associated with impaired cell aggregation and increased migration. To date, the role and underlying molecular mechanisms of STARD8 in GC tissues remain elusive.
In this study, our evidence showed that STARD8, a third member of the deleted liver cancer (DLC) family, may be involved in the progression of gastric carcinomas. Our study found that the expression of STARD8 occurred at very low levels in GC cells and tissues compared with the normal gastric mucosa cell line, GES1, and adjacent normal tissues. The expression of STARD8 in GES1 was higher than in the six different types of GC cell lines and was lowest in HGC27 cells. Furthermore, the expression of STARD8 was not found in 28 cases of GC tissues, whereas all the nonneoplastic tissues expressed STARD8. STARD8 was strongly stained in 51 out of 70 (72.9%) nonneoplastic tissues, but only 11 (15.7%) of GC tissues demonstrated strong staining for STARD8. These results were in agreement with the study that found that STARD8 may be a candidate suppressor gene.14
We further evaluated the correlation between the expression level of STARD8 and clinicopathological parameters of GC. Results showed that the expression level of STARD8 was statistically significantly correlated with differentiation, TNM type and lymph node metastasis. We found that the expression of STARD8 was absent in less-differentiated tissues, weakly expressed in moderately differentiated GCs and strongly expressed in well-differentiated GCs and normal gastric mucosa tissues. Furthermore, compared with TNM stage III and IV samples, TNM stage I and II samples expressed STARD8 more frequently. Patients who had no lymph node metastasis expressed higher levels of STARD8 than those with lymph node metastasis. Previous studies have shown that STARD8 was predominantly localized within intracellular focal adhesion and co-localized with vinculin.14 Results of immunofluorescence analyses showed that the STARD8 protein was expressed in the nuclei and cytoplasm of gastric cells, but more strongly expressed in nuclei. This is in agreement with the previous study and correlated with the STARD8 Rho-GTP activation protein function.
Conclusion
Results of our study suggest that STARD8 may be a tumor suppressor gene and may be associated with biological aggressiveness and poor prognosis in GC. Therefore, further in vitro and in vivo experiments are warranted to elucidate the underlying mechanism of STARD8 in GC and how each type of Rho-GAP interacts in GC.
Acknowledgment
This work was partially supported by grants from the National Natural Science Foundation of China (#30572162), Higher Specialized Research Fund for Doctoral Program of Ministry of Education of China (#20102104110001) and the Natural Science Foundation of Liaoning Province (#201602817).
Disclosure
The authors report no conflicts of interest in this work.
References
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 2016;17(8):496–510. | ||
Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6919):629–635. | ||
Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. | ||
Ellenbroek SI, Collard JG. Rho GTPases: functions and association with cancer. Clin Exp Metastasis. 2007;24(8):657–672. | ||
Kim C, Yang H, Park I, et al. Rho GTPase RhoJ is associated with gastric cancer progression and metastasis. J Cancer. 2016;7(11):1550–1556. | ||
Lukasik D, Wilczek E, Wasiutynski A, Gornicka B. Deleted in liver cancer protein family in human malignancies (Review). Oncol Lett. 2011;2(5):763–768. | ||
Durkin ME, Yuan B-Z, Zhou X, et al. DLC-1: a Rho GTPase-activating protein and tumour suppressor. J Cell Mol Med. 2007;11(5):1185–1207. | ||
Du X, Qian X, Papageorge A, et al. Functional interaction of tumor suppressor DLC1 and caveolin-1 in cancer cells. Cancer Res. 2012;72(17):4405–4416. | ||
Vitiello E, Ferreira JG, Maiato H, Balda MS, Matter K. The tumour suppressor DLC2 ensures mitotic fidelity by coordinating spindle positioning and cell–cell adhesion. Nat Commun. 2014;5:5826. | ||
El-Sitt S, Khalil BD, Hanna S, El-Sabban M, Fakhreddine N, El-Sibai M. DLC2/StarD13 plays a role of a tumor suppressor in astrocytoma. Oncol Rep. 2012;28(2):511–518. | ||
Durkin ME, Ullmannova V, Guan M, Popescu NC. Deleted in liver cancer 3(DLC-3), a novel Rho GTPase-activating protein, is downregulated in cancer and inhibits tumor cell growth. Oncogene. 2007;26(31):4580–4589. | ||
Kawai K, Kiyota M, Seike J, Deki Y, Yagisawa H. START-GAP3/DLC3 is a GAP for RhoA and Cdc42 and is localized in focal adhesions regulating cell morphology. Biochem Biophys Res Commun. 2007;364(4):783–789. | ||
Sun B, Chu D, Li W, et al. Decreased expression of NDRG1 in glioma is related to tumor progression and survival of patients. J Neurooncol. 2009;94(2):213–219. | ||
Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99(2):67–86. | ||
Lehman HL, Van Laere SJ, van Golen CM, Vermeulen PB, Dirix LY, van Golen KL. Regulation of inflammatory breast cancer cell invasion through Akt1/PKBα phosphorylation of RhoCGTPase. Mol Cancer Res. 2012;10(10):1306–1318. | ||
Qian X, Li G, Asmussen HK, et al. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc Natl Acad Sci U S A. 2007;104(21):9012–9017. | ||
Feng X, Li C, Liu W, et al. DLC-1, a candidate tumor suppressor gene, inhibits the proliferation, migration and tumorigenicity of human nasopharyngeal carcinoma cells. Int J Oncol. 2013;42(6):1973–1984. | ||
Jiang Y, Li JM, Luo HQ. Clinicopathological significance of DLC-1 expression in cancer: a meta-analysis. Asian Pac J Cancer Prev. 2015;16(16):7255–7260. | ||
Song LJ, Liu Q, Meng XR, et al. DLC-1 is an independent prognostic marker and potential therapeutic target in hepatocellular cancer. Diagn Pathol. 2016;11:19. | ||
Leung TH, Yam JW, Chan LK, Ching YP, Ng IO. Deleted in liver cancer 2 suppresses cell growth via the regulation of the Raf-1-ERK1/2-p70S6K signalling pathway. Liver Int. 2010;30(9):1315–1323. | ||
Gao K, Li X, Hu G, Yang K, Tian B, Zhang Y. Expression of deleted in liver cancer 2 in colorectal cancer and its correlation with clinicopathological parameters. Oncol Lett. 2012;4(5):988–992. | ||
Mokarram P, Kumar K, Brim H, et al. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 2009;4(9):e7012. | ||
Holeiter G, Bischoff A, Braun AC, et al. The RhoGAP protein Deleted in Liver Cancer 3 (DLC3) is essential for adherens junctions integrity. Oncogenesis. 2012;1:e13. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.