Back to Journals » OncoTargets and Therapy » Volume 12
Downregulation of MMSET impairs breast cancer proliferation and metastasis through inhibiting Wnt/β-catenin signaling
Authors Zhao X, Xie T, Zhao W, Cai W, Su X
Received 29 November 2018
Accepted for publication 11 February 2019
Published 14 March 2019 Volume 2019:12 Pages 1965—1977
DOI https://doi.org/10.2147/OTT.S196430
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Xiaohui Zhao,1,* Tian Xie,1,* Wenhui Zhao,2,3,* Wanhua Cai,1 Xiaobo Su1
1GMU-GIBH Joint School of Life Sciences, Guangzhou Medical University, Guangzhou 511436, China; 2State Key Laboratory of Oncology in Southern China, Sun Yat-sen University, Cancer Center, Guangzhou 510060, China; 3Juancheng People’s Hospital, Juancheng 274600, China
*These authors contributed equally to this work
Background: Recently, the biggest challenge in the treatment of breast cancer is the metastasis of breast cancer cells. Multiple myeloma SET protein (MMSET), a histone lysine methyltransferase, overexpressed in various human cancers, was reported to be associated with carcinogenesis of human cancers.
Methods: Expression of MMSET in breast cancer cell lines and tissues was quantified by real-time PCR and Western blotting. Immunohistochemistry was employed to analyze MMSET expression in 163 clinicopathologically characterized breast cancer cases. Cell functional assays such as MTT assay, colony formation, BrdU assay, flow cytometry, wound healing, Transwell assay, and 3D culture were used to investigate the effect of MMSET in the development and metastasis of human breast cancer. Effects of MMSET on Wnt/β-catenin signaling pathway were further studied by using Western blotting analysis.
Results: Our results showed that MMSET expression was markedly overexpressed in breast cancer cells and clinical specimens and was significantly correlated with patients’ clinicopathologic characteristics and prognosis. Moreover, silencing endogenous MMSET significantly inhibited the proliferation, migration, and metastasis of breast cancer cells through inhibiting the Wnt/β-catenin pathway.
Conclusion: This study found that the downregulated expression of MMSET impaired proliferation and metastasis of human breast cancer through inhibiting Wnt/β-catenin signaling pathway. Notably, our results indicated that MMSET could be a useful biomarker for the prognosis of breast cancer.
Keywords: multiple myeloma SET protein, carcinogenesis, patient’s clinicopathologic characteristics, prognosis, biomarker
Introduction
Breast cancer is one of the most common cancers and the main causes of death in women. It is a heterogeneous disease presenting variable histopathological and biological characteristics, different clinical outcomes, and responses to systemic interventions.1,2 Therefore, there is a need for sophisticated classification systems for this disease, which is clinically practical, easily applicable, and widely reproducible. Currently, there are many methods to classify breast cancer, but no “perfect” classification system has yet been defined. Molecular classification reflecting cellular biomarkers has been used for therapy and prognosis which can be detected by immunohistochemistry.3,4 Understanding the role of key biomarkers in carcinogenesis and in diagnosis will be critical for the treatment of breast cancer. Therefore, it is of great clinical value to further understand the use of critical biomarkers for diagnosis and treatment, to find valuable critical markers as well as novel therapeutic strategies.
Multiple myeloma SET protein (MMSET) belongs to a family of nuclear receptor SET domain (NSD) proteins, whose members encoding lysine methyltransferase activity are overexpressed in many cancer cells.5 In recent studies, the specificity of NSD proteins was diverse, Kuo et al26 reported that MMSET significantly catalyzes the methylation of histone H3K36 in Hela cells, and Ezponda et al6 found that overexpression of MMSET predominantly catalyzes the methylation of H3K36me2 and H3K27me3 in prostate cancer, which correlates with changes in the morphology of prostatic epithelial cells and promotes migration and invasion of prostate cancer.7 Furthermore, overexpression of MMSET leads to t(4;14)(p16;q32) chromosomal translocation, which is cumulatively present in ~15% of multiple myeloma patients.8 In addition, MMSET is also overexpressed in many solid tumor tissues including gastric cancer, and hepatocellular carcinoma. Chesi et al found that MMSET overexpression may be involved in tumor progression and poor prognosis of gastric cancer patients.9 Zhou et al reported that overexpression of MMSET is an independent prognostic factor and is correlated with the worst prognosis and shorter overall survival (OS) time in hepatocellular carcinoma.10 Toyokawa et al have shown that MMSET interacts with Wnt/β-catenin signaling pathway to activate the carcinogenesis of lung cancer.11 Yang et al found that MMSET promotes cancer cell proliferation, survival, and tumor growth via a feed-forward loop through activating the NF-κB signaling pathway.12 Therefore, it is possible that the MMSET exerts a role in the proliferation and metastasis of breast cancer by interacting with the Wnt/β-catenin signaling pathway. These reports indicate that MMSET may be involved in pathogenesis and progression of various cancers; however, clinicopathologic features, function, and mechanism of MMSET in human breast cancer remained to be elucidated.
In the present study, we reported for the first time that the expression of MMSET was upregulated in breast cancer cells and tissues and their correlation with clinicopathologic grading. Importantly, our results indicated that the expression of MMSET was correlated with clinical staging, T classification, N classification, and M classification of the disease. Multivariate analysis revealed that MMSET was an independent predictor for poor survival of breast cancer. Furthermore, we also demonstrated that knockdown of MMSET significantly reduced the proliferation and metastasis of breast cancer cells. Mechanistically, downregulation of MMSET remarkably inhibited Wnt/β-catenin signaling pathway. Collectively, these findings suggested that MMSET played a significant role in human breast cancer progression and pathogenesis and might be a potentially promising biomarker for predicting the prognosis of patients with breast cancer.
Materials and methods
Patient information and tissue samples
This study was conducted on a cohort of 163 cases of paraffin-embedded, archived breast cancer samples, which had been histopathologically and clinically diagnosed at the Sun Yat-sen University Cancer Center from 2002 to 2007. Prior patients’ consent and approval from the institutional clinical and clinicopathological classification and stage were defined according to the TNM staging based on American Joint Committee on Cancer criteria. This study was approved by the Institutional Research Ethics Committee of Sun Yat-sen University Cancer Center. All patients were informed of the purpose of tissue collection and gave written informed consent before the use of these clinical materials for research. This research was carried out in accordance with the Declaration of Helsinki.
Cell lines
Breast cancer cell lines, including MDA-MB-415, MCF-7, HCC1954, BT549, ZR-75-30, MDA-MB-231, T47D, and non-tumorigenic epithelial cell line (MCF-10a) were purchased commercially from American Type Culture Collection (ATCC, Manassas, VA, USA) and were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (HyClone) according to the ATCC protocol.
Plasmids and siRNA transfection
The open reading frames of MMSET and β-catenin were cloned into the mammalian expression vector pcDNA 3.1 (Thermo Fisher Scientific), and pcDNA3 vector as the control. The MMSET siRNA sequence used was 5′-AGGGATCGGAAGAGTCTTCAA-3′, and control siRNA sequence used was 5′-UAACGACGCGACGACGUAATT-3′.13 All the siRNAs were purchased from RiboBio Co., Ltd (Guangzhou, China). Transfection of plasmids or siRNA was performed using Lipofectamine 2000 reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
RNA extraction, reverse transcription (RT) PCR, and real-time PCR
Total RNA from cultured cells was extracted using the Trizol reagent (Thermo Fisher Scientific) as per the manufacturer’s instruction. cDNAs were amplified and quantified in ABI Prism 7500 Sequence Detection System (Thermo Fisher Scientific) using dye SYBR Green I (Thermo Fisher Scientific). The primers were selected as the following: MMSET (forward: 5′-GTGGTAGCTGAGCACCCAGAT-3′ and reverse: 5′-CAGAGTCTTCTTCAGCCTGGA-3′); expression data were normalized to the geometric mean of housekeeping gene GAPDH (forward: 5′-ACCACAGTCCATGCCATCAC-3′ and reverse: 5′-TCCACCACCCTG TTGCTGTA-3′) to control the variability in expression levels and calculated as 2-[(Ct of gene) – (Ct of GAPDH)], where Ct represents the threshold cycle for each transcript.
Western blotting
Western blotting was performed according to standard methods as described previously,14 using anti-MMSET antibody (1:1,000; Abcam, Cambridge, UK), anti-p-GSK3β (Ser9) (1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-GSK3β (1:1,000; Cell Signaling), anti-β-catenin (1:1,000; Cell Signaling), anti-Cyclin D1 (1:2,000; Cell Signaling), and anti-MMP9 (1:1,000; Cell Signaling). Anti-α-tubulin (1:5,000; Cell Signaling) was used as the loading control.
MTT cell viability assay
Cells were seeded in 96-well plates at a density of 2×103 cells/well. At each time point, cells were stained with 100 μL sterile MTT dye (0.5 mg/mL; Sigma-Aldrich Co., St Louis, MO, USA) for 4 hours at 37°C, followed by removal of the culture medium and addition of 100 μL of dimethyl sulfoxide (Sigma-Aldrich Co.). The absorbance was measured at 570 nm, with 655 nm as the reference wavelength. Each experiment was performed in triplicates.
Colony formation assay
Cells were plated in six-well plates (5×102 cells per well) and cultured for 10 days. The colonies were stained with 1% crystal violet for 30 seconds after fixation with 4% formaldehyde for 5 minutes. Colonies were counted, and results were shown as the fold change compared to siControl cells.
Bromodeoxyuridine (BrdU) labeling and immunofluorescence
Cells (5×104) were plated on coverslips (Thermo Fisher Scientific). After 24 hours, the cells were incubated with BrdU for 1 hour and stained with anti-BrdU antibody (Upstate, Billerica, MA, USA) according to the manufacturer’s instruction. BrdU-positive cells were counted under a laser scanning microscope (Carl Zeiss Meditech AG, Jena, Germany) in ten randomly chosen fields from three independent samples, and the results were presented as mean ± SD.
Flow cytometry
Briefly, cells were harvested and fixed in 75% ethanol, stained with propidium iodide (50 μg/mL), and analyzed using a CytoFLEX (Beckman Coulter, Brea, CA, USA). The cell cycle distribution was assessed using ModFit.
Wound healing, cell migration assays
Indicated cells were plated to confluence in six-well plates. Streaks were created in the monolayer with a pipette tip. Progression of migration was observed and photographed at 0, 12, and 24 hours after wounding. ImageJ software was used to quantify the wound healing area of the indicated cells. Cell migration assays were conducted as previously reported.15
3D spheroid invasion assay
The cells were trypsinized and indicated cells (5×103) were seeded on 2% Matrigel-coated 24-well plates, the medium was refreshed every other day. Cells forming a 3D spherical structure (spheres) were photographed at 2-day intervals for 10 days.
Statistical analysis
All statistical analyses were carried out using the SPSS 16.0 statistical software packages. The relationship between MMSET expression and clinicopathologic characteristics was analyzed by the chi-squared test. Survival curves were plotted by the Kaplan–Meier method and compared using the log-rank test. Univariate and multivariate analyses for the potential prognostic factors and the OS were conducted using the Cox proportional hazards regression analysis.
Results
MMSET was upregulated in breast cancer and correlated with progression and poor prognosis of breast cancer patients
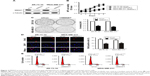
To determine the potential role of MMSET in breast cancer, we first measured MMSET expression, and the results showed that MMSET mRNA and protein expression were markedly upregulated in all the tested breast cancer cell lines compared with non-tumorigenic epithelial cell line (MCF-10a) (Figure 1A and B). Consistently, we found that MMSET expression was higher in six human breast cancer tissues than in their paired adjacent non-tumor tissues (Figures 1C and S1). These results indicated that MMSET expression was upregulated in breast cancer. To investigate the relationship between MMSET expression and the clinicopathological features in breast cancer, the tissue samples of 163 cases of human breast cancer were analyzed by immunohistochemical analysis. These results indicated that MMSET was markedly upregulated in the breast cancer samples (Figure 1D). Statistical analysis further revealed that MMSET expression was strongly associated with the clinical stage (P<0.01) and TNM classification (P<0.01) (Table 1). Kaplan–Meier survival curves and the log-rank test showed that MMSET expression was significantly negatively correlated with the OS of breast cancer patients (P<0.01; Figure 1E). Similar results were obtained from patients in clinical stage I–II and III–IV subgroups (Figure S2). Furthermore, quantitative analysis indicated that the MODs of MMSET staining in clinical stage I–IV primary tumors were significantly higher than those in normal breast tissues (P<0.01; Figure 1F). Moreover, univariate and multivariate analyses indicated that clinical stage, TNM classification, and MMSET expression were independent prognostic factors for the patients (Table 2), suggesting that MMSET might be a predictive biomarker for disease outcome in breast cancer patients.
  | Table 2 Univariate and multivariate analyses of various prognostic parameters in patients with breast cancer |
MMSET promoted the proliferation and regulated the G1–S phase transition of breast cancer cells
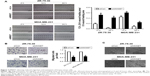
Given its high levels of MMSET expression in breast cancer cells, we first examined the role of MMSET in breast cancer cell proliferation and G1–S phase transition. ZR-75-30 and MDA-MB-231 cells were transfected with siRNAs targeting MMSET. As shown in Figure 2A, MMSET expression was markedly downregulated after siMMSET transfection. MTT and colony formation assays indicated that the proliferation of these two breast cancer cell lines was dramatically inhibited after siMMSET transfection (Figure 2B and C). To further examine the mechanisms of MMSET in stimulation of breast cancer cell proliferation, BrdU incorporation and flow cytometry assays were performed. As shown in Figure 2D and E, silencing MMSET reduced the percentage of S phase cells. In conclusion, our results indicated that MMSET played a critical role in the proliferation and G1–S cell cycle transition of breast cancer cells.
MMSET silencing reduced migration and metastasis of breast cancer cells
To determine the migratory ability of breast cancer cells, wound healing assay was performed and results were shown in Figure 3A. The migration rate of ZR-75-30 and MDA-MB-231 cells at 12 and 24 hours post-scratching was significantly restrained by siMMSET transfection. In addition, Transwell assay without matrigel showed that MMSET silencing drastically inhibited the migration of breast cancer cell lines (Figure 3B). Furthermore, in 3D culture, siMMSET-transduced breast cancer cells grew into less invasive projections emanating from the cells, compared with the siControl cells (Figure 3C). Collectively, our data suggested that MMSET greatly contributes to the development of breast cancer migration and metastasis.
Overexpression of MMSET promoted the proliferation and migration of breast cancer cells
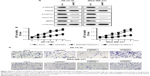
To further confirm the role of MMSET in the proliferation and migration of breast cancer cells, ZR-75-30 and MDA-MB-231 cells were co-transfected with MMSET-siRNA and either pcDNA3.1 (Vector) or MMSET, which encoded the full-length coding sequence of MMSET without its 3′-UTR (Figure 4A). Overexpression of MMSET abrogated the suppressive effects of MMSET-siRNA on the proliferation (Figure 4B and C; P<0.01) and migration (Figure 4D and E; P<0.01) of breast cancer cells. Taken together, these results demonstrate that MMSET is involved in the proliferation and migration of breast cancer cells.
Effects of silenced MMSET expression on Wnt/β-catenin signaling pathway in breast cancer
Since Wnt/β-catenin signaling pathway participates in the proliferation, migration, and metastasis of cancer cells,16 we further focused on the role of MMSET in the regulation of Wnt/β-catenin signaling pathway. Western blotting assay showed that the protein level of p-GSK3β and β-catenin level were strikingly reduced after MMSET silencing in breast cancer cells (Figure 5A). Moreover, the protein levels of cyclinD1 and MMP9, downstream genes of Wnt/β-catenin pathway, were also significantly decreased by knockdown of MMSET in breast cancer cells. To demonstrate the roles of Wnt/β-catenin signaling, β-catenin was forced expressed in MMSET silencing breast cancer cells, and the malignancies were determined. As presented in Figure 5B, the decrease in MMSET-silenced cells’ viability was reversed by the overexpression of β-catenin in breast cancer cells. In addition, transwell assays without matrigel results showed that MMSET siRNA–transduced decline of migratory ability of breast cancer cells was abolished by β-catenin overexpression (Figure 5C). In summary, our results dedicated that downregulation of MMSET impairs breast cancer proliferation and metastasis through inhibiting Wnt/β-catenin signaling.
Discussion
In the present study, we first confirmed that MMSET was remarkably upregulated in breast cancer cell lines and tissues, and it was significantly associated with aggressive stages and unfavorable prognosis of breast cancer patients. By Kaplan–Meier survival curve analysis, we found that the OS time of patients with MMSET high expression was significantly shorter than that of MMSET low expression patients. In particular, the expression of MMSET was significantly associated with shorter OS of patients with stage III/IV breast cancer. Furthermore, we found that MMSET could be an independent prognostic factor for the OS of breast cancer patients, indicated by multivariate Cox regression analysis. Our study suggested that overexpression of MMSET was a common feature in breast cancer and might represent a novel predictive marker for the clinical outcome of the disease.
Epigenetic mechanisms are related to various pathologies such as cancer, cardiovascular diseases, and autoimmune diseases.17 The methylation of histone lysine plays a pivotal role in epigenetic regulation and human carcinogenesis, and the mechanisms need to be investigated further. Studies have shown that the NSD family of HMTases are oncogenes aberrantly expressed in various cancers, suggesting their potential function to serve as novel therapeutic targets.18 The oncogenic role of MMSET was first reported in multiple myeloma and found that MMSET was overexpressed and was associated with poor prognosis of multiple myeloma.19 Recently, numerous studies indicated that MMSET was overexpressed, amplified, or somatically mutated in multiple types of cancer, suggesting its critical role in the development of cancer.20 In this study, we demonstrated that MMSET was both necessary and sufficient to promote proliferation, migration, and metastasis of breast cancer progression. In combination with prior findings, our data supported the hypothesis that MMSET plays an important role in tumorigenesis and metastasis in multiple tumor types.
Wnt/β-catenin signaling pathway has been recognized to take part in tumorigenesis in osteoblasts.21 The β-catenin could be kept in cytoplasm at low level via the enzyme GSK3β-mediated destruction, in the absence of Wnt protein.22 However, when Wnt binds to its receptor, β-catenin escapes from degradation and accumulates in cytoplasm and then enters into nucleus to activate target genes, such as cyclin D1 and MMP9.23 The activation of Wnt/β-catenin signaling pathway confers carcinogenesis and metastasis of multiple tumors.24 Wnt/β-catenin signaling is activated in breast cancer, and the inhibition of Wnt/β-catenin signaling suppresses the proliferation of breast cancer cells.25 In this study, the expression levels of p-GSK3β, β-catenin, and the target protein cyclin D1 and MMP9 were significantly decreased by the downregulation of MMSET in breast cancer cells, and the MMSET-reduced malignant phenotypes were regained after the overexpression of β-catenin.
In conclusion, our study demonstrated that downregulation of MMSET could impair the proliferation and metastasis of breast cancer through inhibiting Wnt/β-catenin signaling pathway. In addition, our results suggest a potential role for MMSET as a clinically independent risk prognostic factor of disease progression, prognosis, and survival in breast cancer patients. Therefore, it is worthwhile to assess the molecular diagnostic ability of MMSET in breast cancer.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (Grant No 81502580), the Science and Technology Department of Guangdong Province (Grant No 201607010014), and Guangzhou Medical University High-level University Academic Key Cultivation Program (Grant No B185004137).
Disclosure
The authors report no conflicts of interest in this work.
References
Watanabe J, Hayashi T, Tadokoro Y, Nishimura S, Takahashi K. Clinical pattern of primary systemic therapy and outcomes of estrogen receptor-positive, HER2-negative metastatic breast cancer: a review of a single institution. Breast Cancer Res Treat. 2017;166(3):911–917. | ||
Nakhlis F, Regan MM, Warren LE, et al. The impact of residual disease after preoperative systemic therapy on clinical outcomes in patients with inflammatory breast cancer. Ann Surg Oncol. 2017;24(9):2563–2569. | ||
Arsenescu R, Bruno MEC, Rogier EW, et al. Signature biomarkers in Crohn’s disease: toward a molecular classification. Mucosal Immunol. 2008;1(5):399–411. | ||
Sideris M, Papagrigoriadis S. Molecular biomarkers and classification models in the evaluation of the prognosis of colorectal cancer. Anticancer Res. 2014;34(5):2061–2068. | ||
Kassambara A, Klein B, Moreaux J. MMSET is overexpressed in cancers: link with tumor aggressiveness. Biochem Biophys Res Commun. 2009;379(4):840–845. | ||
Ezponda T, Popovic R, Shah MY, et al. The histone methyltransferase MMSET/WHSC1 activates Twist1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene. 2013;32(23):2882–2890. | ||
White-Al Habeeb NMA, Garcia J, Fleshner N, Bapat B. Metformin elicits antitumor effects and downregulates the histone methyltransferase multiple myeloma SET domain (MMSET) in prostate cancer cells. Prostate. 2016;76(16):1507–1518. | ||
Keats JJ, Maxwell CA, Taylor BJ, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105(10):4060–4069. | ||
Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92(9):3025–3034. | ||
Zhou P, Wu LL, Wu KM, et al. Overexpression of MMSET is correlation with poor prognosis in hepatocellular carcinoma. Pathol Oncol Res. 2013;19(2):303–309. | ||
Toyokawa G, Cho HS, Masuda K, et al. Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia. 2011;13(10):887–898. | ||
Yang P, Guo L, Duan ZJ, et al. Histone methyltransferase NSD2/MMSET mediates constitutive NF-κB signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol Cell Biol. 2012;32(15):3121–3131. | ||
Li Y, Trojer P, Xu CF, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284(49):34283–34295. | ||
Song L, Li W, Zhang H, et al. Over-expression of AEG-1 significantly associates with tumour aggressiveness and poor prognosis in human non-small cell lung cancer. J Pathol. 2009;219(3):317–326. | ||
Arnold KM, Opdenaker LM, Flynn D, Sims-Mourtada J. Wound healing and cancer stem cells: inflammation as a driver of treatment resistance in breast cancer. Cancer Growth Metastasis. 2015;8:1–13. | ||
Cai WY, Wei TZ, Luo QC, et al. The Wnt – catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J Cell Sci. 2013;126(13):2877–2889. | ||
Zhang Z, Zhang R. Epigenetics in autoimmune diseases: pathogenesis and prospects for therapy. Autoimmun Rev. 2015;14(10):854–863. | ||
Pei H, Zhang L, Luo K, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470(7332):124–128. | ||
Todoerti K, Ronchetti D, Agnelli L, et al. Transcription repression activity is associated with the type I isoform of the MMSET gene involved in t(4;14) in multiple myeloma. Br J Haematol. 2005;131(2):214–218. | ||
Lauring J, Abukhdeir AM, Konishi H, et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111(2):856–864. | ||
Kode A, Manavalan JS, Mosialou I, et al. Leukaemogenesis induced by an activating β-catenin mutation in osteoblasts. Nature. 2014;506(7487):240–244. | ||
Kastritis E, Murray S, Kyriakou F, et al. Somatic mutations of adenomatous polyposis coli gene and nuclear b-catenin accumulation have prognostic significance in invasive urothelial carcinomas: evidence for Wnt pathway implication. Int J Cancer. 2009;124(1):103–108. | ||
Yang S, Liu Y, Li MY, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16(1):124. | ||
Semba S, Kusumi R, Moriya T, Sasano H. Nuclear accumulation of B-Catenin in human endocrine tumors: association with Ki-67 (MIB-1) proliferative activity. Endocr Pathol. 2000;11(3):243–250. | ||
Nadanaka S, Kinouchi H, Kitagawa H. Chondroitin sulfate-mediated N-cadherin/β-catenin signaling is associated with basal-like breast cancer cell invasion. J Biol Chem. 2018;293(2):444–465. | ||
Kuo AJ, Cheung P, Chen K, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;18;44(4):609–620. |
Supplementary materials
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.