Back to Journals » OncoTargets and Therapy » Volume 11
Downregulation of miR-542-3p promotes cancer metastasis through activating TGF-β/Smad signaling in hepatocellular carcinoma
Authors Zhang T, Liu W , Meng W, Zhao H, Yang Q, Gu SJ, Xiao CC, Jia CC, Fu BS
Received 18 October 2017
Accepted for publication 16 January 2018
Published 5 April 2018 Volume 2018:11 Pages 1929—1939
DOI https://doi.org/10.2147/OTT.S154416
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ingrid Espinoza
Tong Zhang,1–3 Wei Liu,4 Wei Meng,1–3 Hui Zhao,1–3 Qing Yang,1–3 Shi-jie Gu,1–3 Cui-cui Xiao,4 Chang-chang Jia,4 Bin-sheng Fu1–3
1Department of Hepatic Surgery, Liver Transplantation Center, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, People’s Republic of China; 2Organ Transplantation Institute of Sun Yat-sen University, Guangzhou, People’s Republic of China; 3Organ Transplantation Research Center of Guangdong Province, Guangzhou, People’s Republic of China; 4Guangdong Key Laboratory of Liver Disease Research, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, People’s Republic of China
Introduction: Hepatocellular carcinoma (HCC) accounts for more than 90% of primary liver cancer. Although great progress has been made on HCC molecular mechanism and therapy techniques, the prognosis of HCC patient is poor due to high metastasis and recurrence.
Materials and methods: Expression of miR-542-3p was quantified by quantitative real-time PCR (qRT-PCR). The role of miR-542-3p in HCC metastasis was examined using transwell and 3D-culture assay. qRT-PCR, Western blotting and luciferase reporter assay were used to elucidate the mechanisms of miR-542-3p-mediated cancer metastasis.
Results and Conclusion: In the research, we found that miR-542-3p is decreased in HCC cell lines and tissues, and downregulation of miR-542-3p enhances, while upregulation suppresses HCC cell invasion ability. Further assay demonstrated that miR-542-3p can directly target TGF-β1 3' untranslated region (3'UTR) to influence TGF-β/Smad signaling pathway, and suppression of miR-542-3p can hyperactivate TGF-β/Smad pathway and further to promote Epithelial-Mesenchyme Transition (EMT) and induce poor prognosis. Lastly, the clinical correlation analysis illustrated that miR-542-3p is negatively related with the activity of TGF-β1. In summary, our results find that miR-542-3p takes an important role on HCC progression and provide more evidence of microRNAs (miRNAs) for cancer therapy.
Keywords: microRNA, HCC, TGF-β, EMT
Introduction
Many factors can cause liver cancer, such as infection by Hepatitis B virus or Hepatitis C virus, aflatoxin contamination, and heavy alcohol consumption. China has been implementing effective measures to prevent liver cancer for the past decades;1 however, as can be seen from the cancer statistics, liver cancer is the sixth most commonly diagnosed cancer and second leading cause of cancer-related death; in particular, China alone accounts for about 50% of the total number of cases and deaths.2–4 And hepatocellular carcinoma (HCC) accounts for more than 90% of primary liver cancer.5 Although significant progress has been made in the fight against HCC, 5-year survival rate remains unsatisfactory owing to high metastasis and recurrence;6,7 therefore it is imperative to clarify the underlying mechanism of HCC metastasis to prolong patient survival.
miRNAs are a cluster of small noncoding RNAs that are 19–25 nucleotides in length.8 They can base-pair to the 3′UTR of mRNA to regulate a gene’s expression. Computational and experimental analysis indicates that miRNAs can bind to over 60% human coding genes.9 miRNAs can regulate genes involved in biological process such as cell cycle, inflammation, and stress response, and most of them function as oncogenes and suppressor genes in cancer,10 such as miR-542-3p. miR-542-3p has been reported as a suppressor genes in several cancers. For example, Rang et al11 reported that miR-542-3p can directly target the protooncogene PIM1 in melanoma, and its downregulation can enhance melanoma cell migration, invasion, and epithelial–mesenchymal transition (EMT) in vitro and in vivo. Long et al12 demonstrated that miR-542-3p is decreased in colorectal cancer, and thus inhibits colorectal cell apoptosis to promote colorectal cancer proliferation and invasion. Althoff et al13 confirmed that miR-542-3p expression is inversely related with the prognosis of neuroblastoma patients, and further analysis suggested that it directly targets survivin to induce cell apoptosis. All these reports demonstrate that miR-542-3p dysregulation is related with many malignancies. However, the important role and molecular mechanism of miR-542-3p in HCC is still poorly understood.
In this research, we found that miR-542-3p is decreased both in HCC cell lines and HCC tissues, and its downregulation induces cancer metastasis and hyperactivity of TGF-β signaling pathway, thus promoting EMT and cancer progression. Our results suggest that miR-542-3p may be of potential as a therapeutic target to prevent HCC metastasis.
Materials and methods
Patient tissue specimens
Three normal liver tissues and nine HCC tissues were collected at the Third Affiliated Hospital of Sun Yat-sen University between 2009 and 2011. Before collecting the samples, patient’s written informed consent and approval from the Institutional Ethics Committee of The Third Affiliated Hospital, Sun Yat-sen University were obtained.
Cells
The immortalized normal hepatocellular cell line (LO2) and human HCC cell lines, BEL7402, HepG2, Hep3B, and QGY-7703, were purchased from the Type Culture Collection Cell Bank, Chinese Academy of Science Committee (Shanghai, People’s Republic of China); human HCC cell lines, HCCC-9810 and Hub7, were purchased from Institute of Basic Medical Science, Chinese Academy of Medical Science (Beijing, People’s Republic of China), The HCC cell lines MHCC97H with high metastasis ability and MHCC97L with low metastasis ability were obtained from the Shanghai Institute of Cell Biology (Shanghai, People’s Republic of China). The immortalized normal hepatocellular cell line (LO2) and human HCC cell lines (BEL-7402, HCCC-9810, HepG2, Hep3B, Huh7, MHCC97L, MHCC97H, and QGY-7703) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA) and 1% penicillin/streptomycin (HyClone). All cell lines were incubated in a humidified atmosphere with 5% CO2 at 37°C.
RNA extraction and quantitative real-time PCR (qRT-PCR)
The total RNA from cells or tissues was extracted with reagent TRIzol (Invitrogen) according to the manufacturer’s protocol. After that, complementary DNA was synthesized by TaqMan miRNA reverse transcription (Applied Biosystems, Foster City, CA, USA). The relative expression of miR-542-3p was quantified with miRNA-specific TaqMan miRNA Assay Kit (Applied Biosystems) in the Applied Biosystems 7500 Sequence Detection system. The relative expression level of miRNA-542-3p was calculated following the formula 2−([Ct of miR-542] − [Ct of U6]) after normalization to U6 small nuclear RNA.
Transwell assay
1×104 indicated cells were seeded into the upper chamber coated with Matrigel (BD Bioscience, San Jose, CA, USA) and DMEM (Invitrogen) without FBS was added, and the lower chamber was filled with culture medium supplemented with 10% FBS (HyClone). After incubation for 24 h, the cells inside the upper chamber that did not pass through the membrane were carefully removed with the cotton swabs. Cells that passed through the coated membrane to the lower surface were fixed with 1% paraformaldehyde for 10 min and subsequently stained with hematoxylin for 5 min. They were finally examined under a microscope, and the number of invasive cells was counted.
3D culture
24-well plates were coated with 100 μL 2% Matrigel (BD Biosciences, San Jose, CA, USA). After Matrigel solidified, 1×104 cells were seeded into plates. The medium was refreshed every 2 days; 14 days later, the cells were examined and pictures were taken.
Western blotting
Western blotting was performed according to the protocol described previously.14 The protein concentration was measured using spectrophotometer (BIO-RAD, Hercules, CA, USA); then, protein was separated on SDS-PAGE and transferred to PVDF membranes. Subsequently, the PVDF membranes were probed with antibody against TGF-β1, E-cadherin, Vimentin, Slug, Snail, Smad2/3, or p84 (1:3,500, Abcam, Cambridge, MA, USA), then probed with HRP-labeled rabbit anti-mouse secondary antibody (1:2,000, Abcam). The GAPDH was used as the cytoplasmic protein loading control and p84 as nuclear protein loading control.
Plasmid, transfection, and luciferase report assay
The sequence of TGF-β1 3′UTR complementary to miR-542-3p seed region was amplified from human genomic DNA. Then, the sequence was cloned into pGL3 luciferase reporter vector (Promega, Madison, WI, USA). The plasmid was transfected into MHCC97H and MHCC97L cell lines with Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA). The luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega) under a luminometer (Berthold Detection System, Pforzheim, Germany), and luciferase activity was normalized to Renilla activity.
For depletion of TGF-β1, the human siRNA sequences of TGF-β1 were cloned into pSuper-retro-puro to generate pSuper-retro-TGF-β1-RNAi. The stable cell line was screened with 0.5 μg/mL puromycin for 10 days.
Resource for predicting the target of miRNA
In the study, miRNA target genes were predicted using TargetScan (http://www.targetscan.org/vert_71/) and miRanda (http://www.microrna.org/microrna/home.do).
Statistical analysis
All values were analyzed using SPSS version 20.0 (SPSS, Chicago, IL, USA) and presented as mean ± standard deviation. Statistical differences were calculated using Student’s t-test, and p<0.05 was considered as statistically significant. Each experiment was repeated three times.
Results
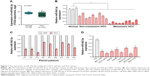
miR-542-3p is decreased in HCC cell lines and tissues
By analyzing the miR-542-3p expression pattern in a public miRNA dataset (The Cancer Genome Atlas, TCGA; https://cancergenome.nih.gov/), we discovered that miR-542-3p is decreased severely in HCC tissues compared to normal liver tissues (normal liver: n=50, HCC: n=372; p<0.001; Figure 1A). Subsequently, qRT-PCR was used to check the miR-542-3p expression level in normal liver tissues, nonmetastatic HCC, and metastatic HCC. The results demonstrated that the expression of miR-542-3p was significantly decreased in HCC tissues compared to normal liver tissues, and the expression in metastatic HCC samples was lower than that in nonmetastatic HCC samples (Figure 1B). The results were also verified in HCC tissues (tumor) and matched adjacent normal tissues. The results showed that miR-542-3p is also markedly decreased in HCC tissues (tumor) compared with matched adjacent normal tissues (Figure 1C). Not only that, we further checked the results in immortalized normal liver epithelial cell line (LO2) and eight HCC cell lines (BEL-7402, HCCC-9810, HepG2, Hep3B, Huh7, MHCC97L, MHCC97H, and QGY-7703). Consistent with the results in tissues, the miR-542-3p expression was significantly inhibited in HCC cell lines compared with the normal cell line (Figure 1D). Altogether, these results illustrate that the expression of miR-542-3p is significantly inhibited in HCC cell lines and HCC tissues.
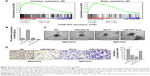
The downregulation of miR-542-3p enhances HCC metastasis
To investigate the role of miR-542-3p in HCC, we analyzed miR-542-3p by gene set enrichment analysis (GSEA) using publicly available HCC patient expression profiles (TCGA; https://cancergenome.nih.gov/) (Figure 2A), and the results demonstrated that miR-542-3p may be involved in cancer metastasis in HCC.
The HCC cell lines MHCC97H with high metastasis ability and MHCC97L with low metastasis ability were used to perform gain-of-function by miR-542-3p mimics (miR-542-3p; RIBOBIO, Guangzhou, People’s Republic of China) or loss-of-function by miR-542-3p inhibitor (miR-542-3p-in; RIBOBIO, Guangzhou, People’s Republic of China). As shown in Figure 2B, the relative expression of miR-542-3p in directed cells was examined by qRT-PCR. 3D-culture assay, which possesses the characteristic of better simulating the in vivo microenvironment, and demonstrated that MHCC97L cells show stronger invasion ability with more outward projection when miR-542-3p is downregulated. However, overexpression of miR-542-3p inhibits the invasion behavior of MHCC97H cells (Figure 2C). In addition, the Transwell assay illustrated that overexpression of miR-542-3p inhibits the migratory and invasive ability compared to the control in MHCC97H; however, inhibition of miR-542-3p enhanced the migratory and invasive ability compared to the control in MHCC97L (Figure 2D). Thus, these results imply that inhibition of miR-542-3p promotes while upregulation suppresses the migration and invasion ability of HCC cell.
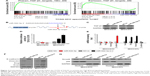
Downregulation of miR-542-3p promotes metastasis via activating the TGF-β signaling pathway in HCC
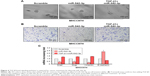
To explore the underlying molecular mechanism by which downregulation of miR-542-3p enhances invasive phenotype in HCC cells, we analyzed the signaling signatures related to miR-542-3p by GSEA in a published HCC dataset (TCGA, https://cancergenome.nih.gov/). The results demonstrated that there may be a negative correlation between miR-542-3p and TGF-β1 signaling pathway (Figure 3A). And not only that, the prediction algorithms TargetScan and miRanda were used to further reveal the precise target by which miR-542-3p inhibits the TGF-β signaling pathway in HCC. The prediction demonstrated that miR-542-3p can directly target the 3′UTR of TGF-β1 (Figure 3B). Next, Western blotting assay suggested that the expression of TGF-β1 is inhibited in MHCC97H cell line when miR-542-3p is overexpressed. However, downregulation of miR-542-3p promotes its expression in MHCC97L cell (Figure 3C). Subsequently, dual luciferase activity assay demonstrated that overexpression of miR-542-3p inhibits and downregulation promotes the luciferase activity of 3′UTR of TGF-β1 in a dose-dependent manner (Figure 3D). EMT is a characteristic of tumor invasion and metastasis, and it is well known that TGF-β1 pathway is involved in EMT to influence cancer progression.15,16 Next, we analyzed the genes involved in EMT, namely, Snail, Slug, Vimentin, and E-cadherin, by RT-PCR and Western blotting. The assay illustrates that overexpression of miR-542-3p robustly decreases while inhibition of miR-542-3p promotes EMT phenotype (Figure 3E and F). Besides, we also detected the expression of TGF-β1 in cell supernatant by enzyme-linked immunosorbent assay and that of the Vimentin and E-cadherin in normal liver tissues, nonmetastatic HCC and metastatic HCC tissues by qPCR, and here also similar trends were observed (Figures S1 and S2). Moreover, Smad2/3, the critical molecular of TGF-β1/Smad signaling pathway, was assessed by Western blotting. As predicted, the miR-542-3p level was inversely relative with the nuclear Smad2/3 expression (Figure 3G). Eventually, TGF-β1 (5 ng/mL; Abcam) can restore the inhibition of invasion phenotype and EMT markers induced by overexpression of miR-542-3p (Figure 4A–C), thus further elucidating that miR-542-3p may promote tumor progression via TGF-β1/Smad signaling pathway. Taken together, our results suggest that downregulation of miR-542-3p promotes metastasis by directly targeting the 3′UTR of TGF-β1 to activate TGF-β1 signaling pathway and induce EMT phenotype in HCC.
The clinical correlation analysis between miR-542-3p and TGF-β1 in HCC tissues
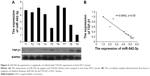
Lastly, we assessed the clinical correlation between miR-542-3p and TGF-β1 in nine fresh HCC tissues. The sample T9 was set as control, as the expression of miR-542-3p was lowest and TGF-β1 the highest. As shown in Figure 5, there is negative correlation between miR-542-3p and TGF-β1 in HCC tissues (r=0.8942, p<0.05; Figure 5).
Altogether, our research shows that miR-542-3p is suppressed and can directly target the 3′UTR of TGF-β1, and suppression of miR-542-3p contributes to EMT phenotype and cancer progression in HCC.
Discussion
In this research, we offered the novel evidence for a link between miR-542-3p and TGF-β/Smad signaling pathway in HCC. The results demonstrate that miR-542-3p is drastically decreased in HCC cell lines and HCC tissues. In addition, overexpression of miR-542-3p inhibits while downregulation miR-542-3p promotes invasive ability. Furthermore, the results imply that downregulation of miR-542-3p activates the TGF-β/Smad signaling pathway via reacting with 3′UTR of TGF-β1. In conclusion, our research indicates that miR-542-3p functions as a suppressor gene and may be a distinctly important target for clinical therapy of HCC.
miR-542-3p has been reported to be a suppressor gene, and its aberrant expression can induce tumor progression and poor prognosis. For example, the study by He et al17 showed that miR-542-3p was decreased and depletion of miR-542-3p enhanced angiogenesis and metastasis in primary breast carcinoma. Althoff et al13 demonstrated that miR-542-3p contributes to suppressing tumor development by directly targeting survivin in neuroblastoma. Moreover, miR-542-3p inhibits cell invasion by decreasing AKT1 phosphorylation and repressing β-catenin nuclear translocation and transactivation activity in human astrocytoma.18 In our research, we found that the downregulation of miR-542-3p can activate TGF-β signaling pathway by directly targeting 3′UTR of TGF-β1. Different researches showed that miR-542-3p can react with different genes, but the above results are not contradictory. On average, computational and experimental analysis manifests that a single miRNA may repress over a hundred mRNA. Not only that, it is predicted that more than 60% human genes contain miRNA-binding sites.10 Such results show that miRNA has powerful functions in biological processes, including occurrence and development of cancer.
In the canonical TGF-β/Smad pathway, active TGF-β1 binds to the cell surface receptor kinases TGF-β1 type I (TβRI) and type II receptors (TβRII), then the TβRII activates TβRI, which further phosphorylates the receptor-associated Smads (R-Smads), Smad2 and Smad3, to regulate gene expression.19 In our research, the amount of Smad2/3 in nuclear changes according to expression of miR-542-3p, such demonstrates that miR-542-3p may function via TGF-β/Smad pathway. Moreover, silencing TGF-β1 reduces the influence of miR-542-3p on TGF-β/Smad pathway, which confirms that TGF-β/Smad pathway is required for miR-452-3p. TGF-β/Smad pathway is vital in EMT.20 Once TGF-β is activated, cells will lose their epithelial phenotype and invade into the surrounding tissues or remote locations, and the EMT downstream effectors will be changed.21,22 For example, the expression of Snail, Slug, and Vimentin will be increased, while that of E-cadherin will be decreased. Herein, we showed that the inhibition of miR-542-3p can activate the TGF-β/Smad pathway in HCC cells, resulting in upregulating Snail, Slug, and Vimentin and downregulating E-cadherin. Loss of E-cadherin causes a series of events to contribute to cell migration and invasion, which then promotes cancer progress. To a certain extent, this explains the phenomenon that miR-542-3p is suppressed in HCC and downregulation of miR-542-3p enhances HCC metastasis.
Conclusion
In summary, this study demonstrates that miR-542-3p is significantly suppressed in HCC cell lines and tissues, which promotes HCC cell invasion ability and cancer progression. Further analysis shows that miR-542-3p can target the 3′UTR of TGF-β1 to activate TGF-β/Smad signaling pathway and promote EMT. Also, there is a negative clinical correlation between miR-542-3p and TGF-β1. The research not only increases the theoretical knowledge but also provides a novel therapeutic strategy for HCC – miR-542-3p. However, there appear to be other deficiencies in this study, and we will perform in vivo experiments in the future research to address these.
Acknowledgment
This work was supported by the Natural Science Foundation of Guangdong Province (grant numbers 2016A030313195, 2014A030313131), the Key Scientific and Technological Projects of Guangdong Province (grant number 2014B020228003, 2014B030301041), and the Science and Technology Planning Project of Guangzhou (grant number 201400000001-3, 158100076).
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27(1):2–12. | ||
Valery PC, Laversanne M, Clark PJ, Petrick JL, McGlynn KA, Bray F. Projections of primary liver cancer to 2030 in 30 countries worldwide. Hepatology. Epub 2017 Aug 31. | ||
Shirvani-Dastgerdi E, Schwartz RE, Ploss A. Hepatocarcinogenesis associated with hepatitis B, delta and C viruses. Curr Opin Virol. 2016;20:1–10. | ||
Gao HJ, Zhao MC, Zhang YJ, et al. Monocarboxylate transporter 4 predicts poor prognosis in hepatocellular carcinoma and is associated with cell proliferation and migration. J Cancer Res Clin Oncol. 2015;141(7):1151–1162. | ||
Ercolani G, Grazi GL, Ravaioli M, et al. Liver resection for hepatocellular carcinoma on cirrhosis: univariate and multivariate analysis of risk factors for intrahepatic recurrence. Ann Surg. 2003;237(4):536–543. | ||
Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struc Mol Biol. 2006;13(12):1097–1101. | ||
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. | ||
Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Ann Rev Pathol. 2014;9:287–314. | ||
Rang Z, Yang G, Wang YW, Cui F. miR-542-3p suppresses invasion and metastasis by targeting the proto-oncogene serine/threonine protein kinase, PIM1, in melanoma. Biochem Biophys Res Commun. 2016;474(2):315–320. | ||
Long HC, Gao X, Lei CJ, et al. miR-542-3p inhibits the growth and invasion of colorectal cancer cells through targeted regulation of cortactin. Int J Mol Med. 2016;37(4):1112–1118. | ||
Althoff K, Lindner S, Odersky A, et al. miR-542-3p exerts tumor suppressive functions in neuroblastoma by downregulating survivin. Int J Cancer. 2015;136(6):1308–1320. | ||
Sun T, Zhao N, Zhao XL, et al. Expression and functional significance of Twist1 in hepatocellular carcinoma: its role in vasculogenic mimicry. Hepatology. 2010;51(2):545–556. | ||
Wang YP, Yu GR, Lee MJ, et al. Lipocalin-2 negatively modulates the epithelial-to-mesenchymal transition in hepatocellular carcinoma through the epidermal growth factor (TGF-beta1)/Lcn2/Twist1 pathway. Hepatology. 2013;58(4):1349–1361. | ||
Sha L, Dong L, Lv L, Bai L, Ji X. HOXB9 promotes epithelial-to-mesenchymal transition via transforming growth factor-beta1 pathway in hepatocellular carcinoma cells. Clin Exp Med. 2015;15(1):55–64. | ||
He T, Qi F, Jia L, et al. MicroRNA-542-3p inhibits tumour angiogenesis by targeting angiopoietin-2. J Pathol. 2014;232(5):499–508. | ||
Cai J, Zhao J, Zhang N, et al. MicroRNA-542-3p suppresses tumor cell invasion via targeting AKT pathway in human astrocytoma. J Biol Chem. 2015;290(41):24678–24688. | ||
Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. | ||
Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–172. | ||
Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-beta targeted cancer therapy. Int J Biol Sci. 2012;8(7):964–978. | ||
Cao S, Cui Y, Xiao H, et al. Upregulation of flotillin-1 promotes invasion and metastasis by activating TGF-beta signaling in nasopharyngeal carcinoma. Oncotarget. 2016;7(4):4252–4264. |
Supplementary materials
  | Figure S1 The TGF-β1 expression is significantly upregulated in cell supernatant, assayed using ELISA. *p<0.05. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.