Back to Journals » OncoTargets and Therapy » Volume 13
Downregulation of Hsa_circ_0000735 Inhibits the Proliferation, Migration, Invasion, and Glycolysis in Non-small-cell Lung Cancer by Targeting miR-940/BMPER Axis
Authors Huang W, Xu X, Liu M, Cui W, Peng G
Received 11 March 2020
Accepted for publication 18 July 2020
Published 24 August 2020 Volume 2020:13 Pages 8427—8439
DOI https://doi.org/10.2147/OTT.S253474
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Carlos E Vigil
Weizhe Huang,1 Xin Xu,2– 5 Mengyang Liu,2– 5 Weixue Cui,2– 5 Guilin Peng2– 5
1Department of Thoracic Surgery, The First Affiliated Hospital of Shantou University Medical College, Shantou 515041, Guangdong, People’s Republic of China; 2Department of Thoracic Surgery, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou 510120, Guangdong, People’s Republic of China; 3State Key Laboratory of Respiratory Disease, Guangzhou 510120, Guangdong, People’s Republic of China; 4National Clinical Research Center for Respiratory Disease, Guangzhou 510120, Guangdong, People’s Republic of China; 5Guangzhou Institute of Respiratory Health, Guangzhou 510120, Guangdong, People’s Republic of China
Correspondence: Guilin Peng Tel +86-20-83062114
Email [email protected]
Background: Lung cancer is the most commonly diagnosed cancer and the major cause of cancer-related deaths worldwide. The increasing studies have demonstrated that circular RNA (circRNA) was involved in the progression of various cancers, including non-small-cell lung cancer (NSCLC). This study was designed to assess the expression, roles and functional mechanisms of circ_0000735 in NSCLC.
Materials and Methods: The expression levels of circ_0000735, miR-940 and bone morphogenetic protein binding endothelial cell precursor-derived regulator (BMPER) were estimated by the real-time quantitative polymerase chain reaction (RT-qPCR). The biological behaviors of NSCLC cells such as proliferation, migration and invasion were analyzed by cell counting kit-8 (CCK-8), colony-forming assays and transwell assay, respectively. Furthermore, extracellular acid ratio and lactate production were tested to assess glycolysis levels of NSCLC cells. The interaction relationship among circ_0000735, BMPER and miR-940 was analyzed by bioinformatics database and dual-luciferase reporter assay. The protein expression level of BMPER was assessed by Western blot assay. Tumorigenesis assay was established to clarify the functional roles of circ_0000735 in vivo.
Results: Circ_0000735 was upregulated and significantly correlated with overall survival in patients with NSCLC. In addition, the loss-of-functional experiments revealed that knockdown of circ_0000735 repressed proliferation, migration, invasion and glycolysis of NSCLC cells and tumor growth in vivo, which was overturned by overexpression of BMPER. Similarly, overexpression of circ_0000735 enhanced proliferation, migration, invasion, and glycolysis of NSCLC cells. In addition, we also confirmed that overexpression of miR-940 impeded proliferation, migration, invasion, and glycolysis of NSCLC cells. Furthermore, overexpression of BMPER abolished si-circ_0000735 induced effects on NSCLC cells.
Conclusion: Circ_0000735 regulated proliferation, migration, invasion, and glycolysis in NSCLC cells by targeting miR-940/BMPER axis.
Keywords: circRNA, circ_0000735, miR-940, BMPER, NSCLC
Introduction
Non-small-cell lung cancer (NSCLC) was the most commonly diagnosed cancer with high incidence and mortality all over the world.1 NSCLC was the main pathological type in clinical practice of lung tumor, which occupied 80–85% of the proportion in all lung cancer cases.2 Currently, poor prognosis of NSCLC might be partly attributed to tumor metastasis and recurrence.3,4 Therefore, research on underlying mechanisms of NSCLC was crucial and urgent.
Increasing evidence has indicated that circular RNA (circRNA), with covalently closed-loop structures without 5ʹ end caps or 3ʹ end poly (A) tails,5 played a key role in carcinogenesis.6 The numerous potential functions to circRNA in human cancer might be attributed to the following factors like enormously abundant, significantly stable, evolutionary conservation, and tissue-specific expression.7 Mechanically, circRNA exerted its biological functions in cellular physiology by various mechanisms including, as miRNA sponge, as intermediate in gene splicing or transcription, interacting with protein, and epigenetic regulation.8,9 Furthermore, Li et al reported that circRNA was a key regulator and novel diagnostic and prognostic biomarker in NSCLC.10 Circ_0000735 was derived from the P2RX1 gene and located on chr17 (3802927–3808661). Li et al revealed that circ_0000735 could target miR-1179/1182 to regulate cell growth, mobility, and apoptosis in NSCLC cells,11 raising questions as to whether circ_0000735 had other target miRNAs in NSCLC.
miRNA was evolutionarily conserved single-stranded RNA with about 21–24 nucleotides in length, which played a critical role in mRNA expression through degradation mRNA or inhibition translation via binding to 3ʹ untranslated region (UTR) of mRNA.12 In addition, numerous studies have reported that miRNA contributed to regulating oncogene expression, which further affected the development of malignant cancers, including NSCLC.13,14 Recent research of miR-940 revealed that miR-940 was a prognostic biomarker in breast cancer,15 papillary thyroid carcinoma,16 and NSCLC.17,18
Furthermore, bone morphogenetic protein binding endothelial cell precursor-derived regulator (BMPER), a secreted glycoprotein, contained five cysteine-rich domains, von Willebrand D domain and a trypsin-inhibitor domain in construction.19 In addition, the bone morphogenetic protein (BMP) signaling pathway was regulated by BMPER in a concentration-dependent fashion.20 Another important thing worth mentioning was that BMP signaling pathway was tightly associated with progression of malignancies.21 Therefore, it was significant to establish the role of circ_0000735 in NSCLC and its relationship with miR-940 and BMPER, which may provide a novel treatment strategy for NSCLC.
Materials and Methods
Patient Specimens
In total 70 paired tissue samples of NSCLC tissue (T) and matched nontumorous tissue (N; at least 3 cm away from the edge of the NSCLC) tissue were received from patients with a surgical procedure at the First Affiliated Hospital of Shantou University Medical College as well as the five-year Kaplan–Meier survival analysis from 2012–2017. In addition, the clinicopathologic features of NSCLC patients were presented in Table 1. The specimens were independently classified with NSCLC by two pathologists. Patients with NSCLC were assigned to different groups based on the median expression levels of circ_0000735 or stage tumor, node, and metastasis (TNM). The human materials were collected with the written informed consent of patients and authorized by the Ethics Committee of the First Affiliated Hospital of Shantou University Medical College. All excised tissue was rapidly transferred to liquid nitrogen and then maintained at −80°C.
 |
Table 1 The Clinical Pathological Characteristics of NSCLC Patients |
Cell Lines and Cell Culture
The human NSCLC cell lines (SK-MES-1, H1299, H1755, A549, and H1581) were obtained from the American Type Culture Collection (Rockville, MD, USA). The human bronchial epithelial cell line (16HBE) was provided form Medical Sciences of Central South University (Hunan, China). Notably, the use of 16HBE was permitted by the Ethics Committee of the First Affiliated Hospital of Shantou University Medical College. All cells were cultured in RPMI 1640 medium (Wisent, Shanghai, China) supplemented with 10% (v/v) fetal bovine serum (FBS; Hyclone, South Logan, UT, USA) and 1% penicillin/streptomycin (Wisent) in a humidified atmosphere containing 5% CO2 at 37°C.
RNA Isolation and Real-time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from tissue and cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) under the manufacturer’s protocols. The complementary DNA was synthesized with 5 μg of total RNA using PrimeScript™ RT reagent kit (Takara, Dalian, China) or Mir-X miR First-Strand Synthesis Kit (Takara). RT-qPCR assay was performed with an SYBR Green Real-Time PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) under AB7300 thermorecycler (Applied Biosystems, Foster City, CA, USA). The parameters of RT-qPCR analysis were consistent with operation of Xu et al.22 The transcript level of circRNA/mRNA or miRNA was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or endogenous small nuclear RNA U6 (U6) based on the 2−ΔΔCt method, respectively. In addition, for analysis of half-life of circ_0000735 and mp2RX1, A549 and H1299 cells were treated with actinomycin D at the indicated time. Moreover, for analysis of location of circ_0000735, RNA in the cytoplasm or nucleus was respectively extracted using NE-PER Reagent (Thermo Fisher Scientific) with GAPDH or U6 as positive controls, correspondingly.
The sequences of primers were displayed:
circ_0000735 (F, 5ʹ-GAAGACAGTGGCTGTACCCC-3ʹ; R, 5ʹ-CGTTGAAGGCCACACACTTG-3ʹ);
p2RX1 (F, 5ʹ-CGTCATCGGGTGGGTGTTTCTCTA-3ʹ; R, 5ʹ-AGGGCGCGGGATGTCGTCA-3ʹ);
miR-940 (F, 5ʹ-GCCGAGAAGGCAGGGCCCCCG-3ʹ; R, 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ);
BMPER (F, 5ʹ-TTTATCACAGACAACCCTTGCAT-3ʹ; R, 5ʹ-TCCTGGCACTGGCGTAGAA-3ʹ);
GAPDH (F, 5ʹ-TCCCATCACCATCTTCCAGG-3ʹ; R, 5ʹ-GATGACCCTTTTGGCTCCC-3ʹ);
U6 (F, 5ʹ-AACGAGACGACGACAGAC-3ʹ; R, 5ʹ-GCAAATTCGTGAAGCGTTCCATA-3ʹ).
Transfection Assay
Specific small interfering RNA (siRNA) objecting circ_0000735 (si- circ_0000735) and siRNA scrambled control (si-NC), miR-940 mimic (miR-940) and its negative control (miR-NC), and miR-940 inhibitor (anti-miR-940) and its negative control (anti-NC) were generated and synthesized by Biossci Company (Wuhan, China). The lentiviral vectors with sh-circ_0000735 (sh-circ_0000735) or matched control (sh-NC), and circ_0000735-overexpression vector (circ_0000735), BMPER-overexpression vector (BMPER) and their negative control (vector) were purchased from RiboBio Corporation (Guangzhou, China). Lipofectamine 2000 (Invitrogen) was utilized for oligonucleotides and vectors transfection as instructed by the manufacturer.
Cell Proliferation Assay
Cell counting kit-8 (CCK-8) assay was conducted to assay cell proliferation in vitro as described by Xue et al.23 A549 and H1299 cells were injected into 96-well plates at a concentration of 1000 cells/well. After incubation for 24 h the 10 μL of CCK-8 reagent (Dojindo Laboratories, Kumamoto, Japan) was added to each well and incubated for four hours in a humidified incubator at 37°C with 5% CO2. At the end of incubation, absorbance value (OD) of each well was measured at 450 nm on a microplate spectrophotometer (BioTek Instruments, Winooski, VT, USA). Furthermore, colony-forming assay was performed as described by Wang et al.24 In brief, stably transfected A549 and H1299 cells (500 cells/well) were seeded into a 12-well plate and then cultured at 37°C with 5% CO2. At the 14 d time points, the A549 and H1299 cells were fixed and stained with 0.5% crystal violet solution. A microscope (Olympus, Tokyo, Japan) was used to record colonies of different groups.
Transwell Migration and Invasion Assays
The 24-well transwell chamber adhered with matrigel (Becton Dickinson, San Jose, CA, USA) or without matrigel was applied to assess cell invasion or migration ability, correspondingly. NSCLC cells (2×104 cells) in 200 μL of serum-free medium were added into the upper chamber, whereas the lower chamber was filled with 600 μL of complete medium containing 10% FBS. After incubation for 24 h, migrated or invaded cells were fastened with 4% formaldehyde and then dyed with 0.1% crystal violet. For transwell assay, a microscope (Olympus, Tokyo, Japan) and Image J software (National Institutes of Health, Bethesda, MD, USA) were used to photograph and calculate cell number of migrated or invaded cells.
Extracellular Acidification Rate (ECAR) and Lactate Production Assays
ECAR assay was carried out using the commercial Seahorse XF Glycolysis Stress Test Kit (Agilent Technologies, Suzhou, China) in the light of the user’s guideline and previously described.25 Furthermore, the Lactate Assay Kit (Solarbio, Beijing, China) was used for detection lactate production based on user’s guideline.
Dual-luciferase Reporter Assay
We predicted the miR-940 binding sites in circ_0000735 or 3ʹUTR of BMPER using the bioinformatics databases circular RNA Interactome (https://circinteractome.nia.nih.gov/bin/mirnasearch) or TargetScanHuman (http://www.targetscan.org/vert_72/), respectively. The fragments of circ_0000735 or BMPER 3ʹ UTR containing the predicted wild and mutant sites on miR-940 were synthesized and directly inserted into PGL3 vector (Promega, Madison, WI, USA) to establish luciferase reporter vectors WT-circ_0000735, MUT-circ_0000735, WT-BMPER 3ʹ UTR, or MUT-BMPER 3ʹ UTR, respectively. A549 and H1299 cells were co-transfected with indicated vectors along with miR-940 mimic or miR-NC. After 48 h of culture, the luciferase activity was detected by the dual-luciferase reporter assay system (Promega), with Renilla luciferase activity as the internal control.
Western Blot Assay
Briefly, cells or tissue were lysed with radio-immunoprecipitation assay (RIPA; Beyotime, Shanghai, China) buffer and protease inhibitors (Beyotime) then centrifuged for 10 min (12,000×g, 4°C). Total protein for each sample was subjected to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and shifted onto polyvinylidene fluoride (GE Healthcare, Piscataway, NJ, USA). Membranes were immersed with 3% albumin bovine V (Amyjet scientific, Wuhan, China) in Tris-buffered saline and then incubated with primary antibody anti-BMPER (1:1000 dilution; ab75183; Abcam, Cambridge, MA, USA) overnight at 4°C, with GAPDH (1:2000 dilution; ab181602; Abcam) as the internal control. Afterwards, the blots were re-probed with secondary antibody conjugated with horseradish peroxidase (1:2000 dilution; ab205718; Abcam). Finally, immunoreactive bands were visualized and analyzed with ECL Western Blotting Detection Kit (Solarbio) and Image J software (National Institutes of Health), respectively.
In vivo Experiment
The animal experiment was strictly obeyed the guidelines of China Science Review of Laboratory Animal Welfare, and this animal experiment was permitted by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Shantou University Medical College. A549 cells stably transfected with lentiviral vectors with sh-circ_0000735 (sh-circ_0000735) or matched control (sh-NC) were hypodermically injected into right flanks of 12 female nude mice (four weeks; Shanghai Experimental Animal Center, Shanghai, China), respectively. After 28 days, the mice were euthanized and the tumors were removed for weight detection and RT-qPCR assay.
Statistical Analysis
All analyses were carried out using the SPSS 21.0 software (IBM, Somers, NY, USA). The differences of results were assessed by using Student’s t-test when there were only two groups, or analyzed by one-way analysis of variance when there were multiple groups. All data were exhibited as mean ±standard deviation and P-value less than 0.05 was considered statistically significant.
Results
Circ_0000735 was Upregulated in NSCLC Tissue and Cells and Connected with Poor Prognosis of NSCLC
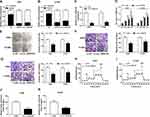
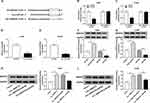
The expression level of circ_0000735 was analyzed using RT-qPCR analysis in cancerous tissue and neighboring normal tissue from NSCLC patients. Circ_0000735 expression was considerably higher in NSCLC tissue relative to contiguous normal tissue (Figure 1A). The 70 NSCLC patients were separated into I+II (n=40) or III+IV (n=30) grade groups in accordance with World Health Organization classification. As shown in Figure 1B, the advanced TNM stage showed higher circ_0000735 level than early group. In accordance with the median expression level of circ_0000735, patients were divided into two groups. We found that cumulative survival time of NSCLC patients with a high level of circ_0000735 was shorter than that with low level of circ_0000735 by Kaplan–Meier curve analysis (Figure 1C). We also measured the expression of circ_0000735 in SK-MES-1, H1299, H1755, A549, H1581, and 16HBE cells, and the results indicated that the expression of circ_0000735 was upregulated in NSCLC cells compared to that in 16HBE cells, especially in A549 and H1299 cells (Figure 1D). Actinomycin D was used to repress transcription in A549 and H1299 cells. As displayed in Figure 1E and F, circ_0000735 was more stable than mp2RX1 by performing RT-qPCR assay. Taken together, circ_0000735 was increased in NSCLC cells and tissue, which might be correlated with poor outcome of NSCLC patients.
Initially, RT-qPCR assay suggested the predominant cytoplasmic distribution of circ_0000735, instead of nuclear (Figure 2A and B). Subsequently, we transfected si-circ_0000735 into A549 and H1299 cells, with si-NC as control. The results of RT-qPCR assay showed that the inhibition of circ_0000735 was found in A549 and H1299 cells after transfection with si-circ_0000735 (Figure 2C). CCK-8 and colony-forming assays indicated that knockdown of circ_0000735 profoundly impeded proliferation ability of NSCLC cells (Figure 2D and E). Consistently, migration and invasion capacities of A549 and H1299 cells declined significantly by silencing of circ_0000735 (Figure 2F and G). The results of the ECAR assay indicated that basal glycolysis and maximum glycolytic capacity declined in A549 and H1299 cells after circ_0000735 knockdown (Figure 2H and I). Knockdown of circ_0000735 led to an obvious decrease of lactate production in A549 and H1299 cells (Figure 2J and K). All the above date implied that suppression of circ_0000735 blocked proliferation, migration, invasion and glycolysis in NSCLC cells.
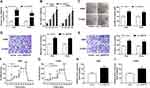
As presented in Figure 3A, circ_0000735 was increased in A549 and H1299 cells after transfection with circ_0000735. Furthermore, overexpression of circ_0000735 enhanced proliferation of A549 and H1299 cells (Figure 3B and C). After overexpression of circ_0000735, A549 and H1299 cells showed higher migration and invasion than the negative group (Figure 3D and E). The results of ECAR assay revealed that basal glycolysis and maximum glycolytic capacity were more enhanced in the circ_0000735 group than the vector group (Figure 3F and G). Consequently, lactate production was increased in A549 and H1299 cells with circ_0000735-overexpressing compared with the control group (Figure 3H and I). Based on all data, circ_0000735 might play an oncogene role in NSCLC.
We found that a supposed circ_0000735 binding site was located in the miR-490 (Figure 4A). The elevated expression of miR-490 noticeably repressed the luciferase activity of the WT-circ_0000735 reporter vector by 70%, whereas luciferase activity of the MUT- circ_0000735 was not changed by the transfection of miR-490 mimic (Figure 4B and C). After that, we used RT-qPCR to assess the effect of circ_0000735 overexpression on miR-490 expression. The results indicated that shortage of circ_0000735 remarkably enhanced miR-490 levels, while the opposite phenomenon was found in A549 and H1299 cells transfected with si-circ_0000735 (Figure 4D and E). In summary, miR-490 was negatively regulated by circ_0000735 in NSCLC.
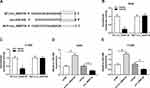
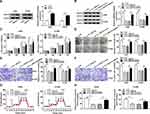
Overexpression of miR-940 Constrained the Growth, Migration, Invasion and Glycolysis of NSCLC Cells
miR-940 was downregulated in NSCLC tissue and cells compared with matched adjacent normal tissue and 16HBE, correspondingly (Figure 5A and B). Furthermore, miR-940 was increased in A549 and H1299 cells after transfection with miR-940 compared with the control (Figure 5C). The cell proliferation was greatly reduced in A549 and H1299 cells by miR-940 overexpression (Figure 5D and E). Analogously, the significant decrease of migration and invasion abilities of A549 and H1299 cells could be seen in the group of miR-940, compared the group of miR-NC (Figure 5F and G). Moreover, the glycolytic capacity and lactate production were reduced by the overexpression of miR-940 in A549 and H1299 cells (Figure 5H and I). In summary, these data indicated an important role for miR-940 in growth, migration, invasion, and glycolysis of NSCLC cells.
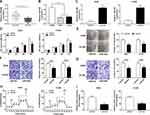
miR-490 Interacted with the 3ʹUTR of BMPER in NSCLC Cells
The miR-490 putative binding sites and matched mutant sites in the 3ʹUTR of BMPER were shown in Figure 6A. To explore whether miR-490 could interact with BMPER, the sequences of BMPER 3ʹUTR containing intact target sites of miR-490 or matched mutation were cloned into luciferase reporter vector. The luciferase activity was greatly inhibited in A549 and H1299 cells co-transfected with miR-490 and WT-BMPER 3ʹUTR, but not that of MUT-BMPER 3ʹUTR (Figure 6B and C). Next, we analyzed the expression level of miR-490 in A549 and H1299 cells transfected with anti-miR-940 or anti-NC. The results suggested that miR-490 expression in anti-miR-940 group was lower than that in anti-NC group (Figure 6D and E). Importantly, BMPER was upregulated in A549 and H1299 cells transfected with anti-miR-940, while miR-940 mimic generated opposite results (Figure 6F and G). Also, A549 and H1299 cells transfected with circ_0000735 had an obvious enhancement of BMPER expression, which was abolished by reintroduction of miR-940 (Figure 6H and I). These data uncovered that circ_0000735 regulated BMPER expression by sponging miR-940.
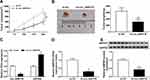
Overexpression of BMPER was validated in A549 and H1299 cells transfected with BMPER overexpression vector by performing Western blot assay (Figure 7A). Moreover, deficiency of circ_0000735 resulted in decreased expression of BMPER, whereas it was rescued by BMPER overexpression (Figure 7B). We also investigated the effect of BMPER overexpression on cell proliferation. The results indicated that upregulation of BMPER could resume si-circ_0000735-mediated repression for proliferation (Figure 7C and D). In addition, the results of transwell analysis displayed that knockdown of circ_0000735 drastically impaired the migration and invasion of A549 and H1299 cells, while this inhibition was attenuated by re-expressing BMPER (Figure 7E and F). The basal glycolysis and maximum glycolytic capacity were declined in A549 and H1299 cells transfected with si-circ_0000735, whereas co-transfection with si-circ_0000735 and BMPER attenuated these effects (Figure 7G). We also found that A549 and H1299 cells transfected with si-circ_0000735 exhibited a lower lactate production level than si-NC group, which was inverted by upregulating BMPER (Figure 7H). In summary, above data supported that circ_0000735 silencing could impede proliferation, migration, invasion and glycolysis of NSCLC cells by affecting BMPER expression.
To assess the role of circ_0000735 in NSCLC progression, A549 cells that stably transfected with lentiviral vectors with sh-circ_0000735 or corresponding control (sh-NC) were inoculated into nude mice. The xenograft experiments confirmed that the downregulation of circ_0000735 obviously suppressed subcutaneous tumor growth and weight compared with the sh-NC control group (Figure 8A and B). Additionally, RT-qPCR analysis revealed that circ_0000735 declined, while miR-940 increased in the sh-circ_0000735 group when compared with the sh-NC group (Figure 8C). In addition, silencing of circ_0000735 also suppressed BMPER expression (Figure 8D and E). Above data suggested that circ_0000735 silenced impeding tumor growth in vivo by regulation of miR-940 and BMPER.
Discussion
Our study revealed that circ_0000735 was upregulated in NSCLC tissue and cells compared with control groups. Simultaneously, the expression level of circ_0000735 was closely connected with poor survival of NSCLC patients. Loss-of functional experiments uncovered that downregulation of circ_0000735 impeded cell growth, mobility, and glucose metabolism of NSCLC cells and tumor growth in vivo. Mechanically, circ_0000735 modulated miR-940 to regulate BMPER expression. Collectively, our findings showed that circ_0000735/miR-940/BMPER axis played an important role in NSCLC progression.
Circ_0000735 was upregulated in NSCLC tissue rather than nontumorous tissue.11,26 In this study, we clarified the mechanism of circ_0000735 in the progression of NSCLC. Based on obvious findings,11 we wondered whether circ_0000735 could adsorb other miRNAs to regulate NSCLC. miR-940 could mediate family sequence similarity 83 member F (FAM83F) expression, consequently repressing the NSCLC cell proliferation.17 In addition, miR-940 could inhibit cell invasion by targeting snail in NSCLC.18 Analogous tumor inhibition impacts by miR-940 were also reported in other types of tumors such as hepatocellular carcinoma and glioblastoma.27,28 Nevertheless, Zhou et al confirmed that miR-940 was upregulated in endometrial carcinoma tissue and cells rather than controls, and its expression was associated with poor outcome.29 The different functions of miR-940 in human cancers may be attributed to tissue-specific expression of miRNA. By searching online database, circular RNA Interactome, miR-940 was identified as a potential target of circ_0000735, and we also confirmed that miR-940 was significantly reduced in NSCLC tissue and cells compared to normal controls. Besides, elevated expression of miR-940 inhibited proliferation, migration, invasion, and glycolysis of NSCLC cells. Importantly, we also demonstrated that BMPER was the functional target of miR-940.
It was well recognized that activation of the BMP pathway was tightly connected with tumor evolution. BMP, extracellular protein, could signal via interacting cell-surface receptors of serine/threonine kinase, consequently activating intracellular signaling.30,31 Importantly, BMP signaling pathway was regulated by multiple factors, including BMPER, and BMPER could directly interact with BMP2,32 BMP4,33 and BMP6.34 Heinke et al reported that BMPER performed a vital function in tumorigenesis by regulating of behavior of malignant cells such as proliferation and migration.35 Importantly, overexpression of BMPER was confirmed as an important phenotype of malignant tumor.36 In addition, Lockyer et al reported the anti-inflammatory function of BMPER, which could greatly relieve pulmonary inflammation and injury of mice caused by lipopolysaccharide.37 Here, we revealed the carcinogenic function of BMPER in NSCLC, overexpression of BMPER overturned si-circ_0000735-induced the effects on proliferation, migration, invasion, and glycolysis in NSCLC cells. Conversely, loss of BMPER in A549 impeded proliferation and invasion in vitro as well as tumorigenesis in vivo, and further pointed out that overexpression of BMPER was a characteristic of malignant tumors.35
Conclusively, our study revealed that circ_0000735 was overexpressed in NSCLC. Importantly, high-expression of circ_0000735 was closely associated with NSCLC development, indicating that circ_0000735 might be a possible biomarker for NSCLC patients, which might be useful in NSCLC diagnosis. After that, functional experiments further suggested that circ_0000735 promoted NSCLC progression via sponging miR-940 to regulate BMPER, thereby regulating proliferation, migration, invasion, and glycolysis of NSCLC cells. Furthermore, the circ_0000735/miR-940/BMPER signaling pathway may be a potential diagnosis target for NSCLC. In the current study, our results extended the role of circ_0000735 and enriched data basis for NSCLC diagnosis, which might be helpful in developing potential targets in NSCLC therapies.
Conclusion
Collectively, our data suggested that circ_0000735 may be a prognostic biomarker in patients with NSCLC. The oncogenic function of circ_0000735 was attributed to its regulation of miR-940/BMPER axis in NSCLC. Moreover, the regulatory role of the circ_0000735/miR-940/BMPER pathway in NSCLC was preliminarily confirmed.
Highlights
- Circ_0000735 was elevated in NSCLC tissue and cells and was associated with poor prognosis.
- Knockdown of circ_0000735 impeded proliferation, migration and invasion of NSCLC cells.
- Circ_0000735 regulated BMPER by sponging miR-940 was first reported.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Wu YL, Planchard D, Lu S, et al. Pan-Asian adapted Clinical Practice Guidelines for the management of patients with metastatic non-small-cell lung cancer: a CSCO-ESMO initiative endorsed by JSMO, KSMO, MOS, SSO and TOS. Ann Oncol. 2019;30(2):171–210. doi:10.1093/annonc/mdy554
3. Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi:10.1016/j.cell.2006.11.001
4. Uramoto H, Tanaka F. Recurrence after surgery in patients with NSCLC. Transl Lung Cancer Res. 2014;3(4):242–249.
5. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440–441. doi:10.1126/science.1238522
6. Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi:10.1016/j.canlet.2018.03.035
7. Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5(2):472–480.
8. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi:10.1261/rna.047126.114
9. Zhao X, Cai Y, Xu J. Circular RNAs: biogenesis, mechanism, and function in human cancers. Int J Mol Sci. 2019;20(16):3926. doi:10.3390/ijms20163926
10. Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145(12):2875–2889. doi:10.1007/s00432-019-03045-4
11. Li W, Jiang W, Liu T, et al. Enhanced expression of circ_0000735 forecasts clinical severity in NSCLC and promotes cell progression via sponging miR-1179 and miR-1182. Biochem Biophys Res Commun. 2019;510(3):467–471. doi:10.1016/j.bbrc.2019.01.134
12. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
13. Li Q, Wu X, Guo L, et al. MicroRNA-7-5p induces cell growth inhibition, cell cycle arrest and apoptosis by targeting PAK2 in non-small cell lung cancer. FEBS Open Biol. 2019;9(11):1983–1993. doi:10.1002/2211-5463.12738
14. Wang Z, Han Z, Zhang L, et al. MicroRNA-98-5p regulates the proliferation and apoptosis of A549 cells by targeting MAP4K3. Oncol Lett. 2019;18(4):4288–4293.
15. Liu W, Xu Y, Guan H, et al. Clinical potential of miR-940 as a diagnostic and prognostic biomarker in breast cancer patients. Cancer Biomark. 2018;22(3):487–493. doi:10.3233/CBM-171124
16. Hu J, Li C, Liu C, et al. Expressions of miRNAs in papillary thyroid carcinoma and their associations with the clinical characteristics of PTC. Cancer Biomark. 2017;18(1):87–94. doi:10.3233/CBM-161723
17. Gu GM, Zhan YY, Abuduwaili K, et al. MiR-940 inhibits the progression of NSCLC by targeting FAM83F. Eur Rev Med Pharmacol Sci. 2018;22(18):5964–5971.
18. Jiang K, Zhao T, Shen M, et al. MiR-940 inhibits TGF-beta-induced epithelial-mesenchymal transition and cell invasion by targeting Snail in non-small cell lung cancer. J Cancer. 2019;10(12):2735–2744. doi:10.7150/jca.31800
19. Moser M, Binder O, Wu Y, et al. BMPER, a novel endothelial cell precursor-derived protein, antagonizes bone morphogenetic protein signaling and endothelial cell differentiation. Mol Cell Biol. 2003;23(16):5664–5679. doi:10.1128/MCB.23.16.5664-5679.2003
20. Heinke J, Wehofsits L, Zhou Q, et al. BMPER is an endothelial cell regulator and controls bone morphogenetic protein-4-dependent angiogenesis. Circ Res. 2008;103(8):804–812. doi:10.1161/CIRCRESAHA.108.178434
21. Dituri F, Cossu C, Mancarella S, et al. The interactivity between TGFbeta and BMP signaling in organogenesis, fibrosis, and cancer. Cells. 2019;8(10):1130. doi:10.3390/cells8101130
22. Xu Y, Wang J, Qiu M, et al. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biol. 2015;36(3):1643–1651. doi:10.1007/s13277-014-2763-6
23. Xue M, Shi D, Xu G, Wang W. The long noncoding RNA linc00858 promotes progress of lung cancer through miR-3182/MMP2 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):2091–2097. doi:10.1080/21691401.2019.1617728
24. Wang X, Qi G, Zhang J, et al. Knockdown of long noncoding RNA small nucleolar RNA host gene 12 inhibits cell growth and induces apoptosis by upregulating miR-138 in nonsmall cell lung cancer. DNA Cell Biol. 2017;36(11):892–900. doi:10.1089/dna.2017.3830
25. Zhao SJ, Shen YF, Li Q, et al. SLIT2/ROBO1 axis contributes to the Warburg effect in osteosarcoma through activation of SRC/ERK/c-MYC/PFKFB2 pathway. Cell Death Dis. 2018;9(3):390. doi:10.1038/s41419-018-0419-y
26. Zhang S, Zeng X, Ding T, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8(1):2878. doi:10.1038/s41598-018-21300-5
27. Li P, Xiao Z, Luo J, et al. MiR-139-5p, miR-940 and miR-193a-5p inhibit the growth of hepatocellular carcinoma by targeting SPOCK1. J Cell Mol Med. 2019;23(4):2475–2488. doi:10.1111/jcmm.14121
28. Luo H, Xu R, Chen B, et al. MicroRNA-940 inhibits glioma cells proliferation and cell cycle progression by targeting CKS1. Am J Transl Res. 2019;11(8):4851–4865.
29. Zhou Z, Xu YP, Wang LJ, et al. miR-940 potentially promotes proliferation and metastasis of endometrial carcinoma through regulation of MRVI1. Biosci Rep. 2019;39(6):2. doi:10.1042/BSR20190077
30. Moser M, Patterson C. Bone morphogenetic proteins and vascular differentiation: bMPing up vasculogenesis. Thromb Haemost. 2005;94(4):713–718.
31. Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8(12):970–982. doi:10.1038/nrm2297
32. Dyer L, Lockyer P, Wu Y, et al. BMPER promotes epithelial-mesenchymal transition in the developing cardiac cushions. PLoS One. 2015;10(9):e0139209. doi:10.1371/journal.pone.0139209
33. Lockhart-Cairns MP, Lim KTW, Zuk A, et al. Internal cleavage and synergy with twisted gastrulation enhance BMP inhibition by BMPER. Matrix Biol. 2019;77:73–86. doi:10.1016/j.matbio.2018.08.006
34. Patel N, Masaratana P, Diaz-Castro J, et al. BMPER protein is a negative regulator of hepcidin and is up-regulated in hypotransferrinemic mice. J Biol Chem. 2012;287(6):4099–4106. doi:10.1074/jbc.M111.310789
35. Heinke J, Kerber M, Rahner S, et al. Bone morphogenetic protein modulator BMPER is highly expressed in malignant tumors and controls invasive cell behavior. Oncogene. 2012;31(24):2919–2930. doi:10.1038/onc.2011.473
36. Lin KY, Lu D, Hung CF, et al. Ectopic expression of vascular cell adhesion molecule-1 as a new mechanism for tumor immune evasion. Cancer Res. 2007;67(4):1832–1841. doi:10.1158/0008-5472.CAN-06-3014
37. Lockyer P, Mao H, Fan Q, et al. LRP1-dependent BMPER signaling regulates lipopolysaccharide-induced vascular inflammation. Arterioscler Thromb Vasc Biol. 2017;37(8):1524–1535. doi:10.1161/ATVBAHA.117.309521
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.