Back to Journals » Cancer Management and Research » Volume 12
Downregulation of circRNA_0000285 Suppresses Cervical Cancer Development by Regulating miR197-3p–ELK1 Axis
Received 9 March 2020
Accepted for publication 16 July 2020
Published 18 September 2020 Volume 2020:12 Pages 8663—8674
DOI https://doi.org/10.2147/CMAR.S253174
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Wenmin Zhang,1 Suping Zhang2
1Department of Obstetrics and Gynecology, Heze Municipal Hospital, Heze, Shandong 274000, People’s Republic of China; 2Department of Reproductive, Zoucheng People’s Hospital, Zoucheng, Shandong 273500, People’s Republic of China
Correspondence: Suping Zhang Tel +86-537-6626135
Email [email protected]
Background: Circular RNAs (circRNAs) are involved in the development of human cancers, including cervical cancer (CC). However, the role and mechanism of the circRNA hsa_circ_0000285 (circ_0000285) in CC development remain largely unknown.
Methods: Thirty paired CC and adjacent normal tissue samples were harvested. CC cell lines SiHa and HeLa were cultured in this study. The expression of circ_0000285, miR197-3p and ELK1 was detected via qRT-PCR or Western blot. CC development was assessed via cell viability, colony formation, apoptosis, cell cycle, and autophagy using MTT, colony-formation assays, flow cytometry and Western blot. The target association was analyzed via dual luciferase–reporter assay, RNA immunoprecipitation, and RNA pull-down. The role of circ_0000285 in CC in vivo was analyzed using a xenograft model.
Results: circ_0000285 abundance was enhanced in CC tissue and cells and mainly located in cytoplasm. Silence of circ_0000285 suppressed cell viability and colony formation, arrested the cell cycle at the G0/G1 phase, and induced apoptosis and autophagy in CC cells. miR197-3p was targeted by circ_0000285, and miR197-3p knockdown reversed the effect of circ_0000285 silence on CC development. miR197-3p directly targeted ELK1 to inhibit CC development. circ_0000285 regulated ELK1 by modulating miR197-3p. Knockdown of circ_0000285 reduced xenograft tumor growth in vivo.
Conclusion: Knockdown of circ_0000285 repressed CC development by increasing miR197-3p and decreasing ELK1.
Keywords: cervical cancer, circ_0000285, miR197-3p, ELK1
Introduction
Cervical cancer (CC) is a common disorder of female reproductive system, including squamous-cell carcinoma and adenocarcinoma.1 It is one of the leading causes of cancer-related death in females, with high incidence and mortality worldwide.2 Current options for CC treatment have made considerable advances, while outcomes for CC patients at the advanced stage remain poor.3 As such, new insights into the pathogenesis and treatment of CC are urgently needed.
Recently, a study focused on the role of circular RNAs (circRNAs) in gynecological diseases, including CC.4 circRNAs are a group of noncoding RNAs produced via covalently closed loops through back-splicing with a lack of polyadenylation and capping.5 circRNAs play important roles in the development, diagnosis, prognosis, and treatment of human cancers.6 They may regulate miRNAs and their targeted genes through the competing endogenous RNA (ceRNA) network.7 Moreover, dysregulated circRNAs are implicated in the carcinogenesis and development of CC.8 hsa_circ_0000285 (circ_0000285) is a circRNA derived from HIPK3, which has an important role in CC development.9 Studies have indicated that circ_0000285 takes part in malignancies of multiple cancers, including nasopharyngeal carcinoma, bladder cancer, osteosarcoma, and laryngocarcinoma.10,13 More importantly, emerging evidence suggests that circ_0000285 promotes proliferation and metastasis of CC.14 Although dysregulated circ_0000285 is involved in CC, how circ_0000285 regulates CC development is largely unclear.
miRNAs are noncoding RNA molecules with 19–25 nucleotides that exhibit key roles in the development and treatment of CC.15 miR197-3p has been suggested to participate in cancer development via serving as an oncogene or tumor suppressor.16,17 Moreover, miR197-3p can suppress proliferation and invasion of CC cells.18 However, whether miR197-3p is required for circ_0000285-mediated regulation of CC development is unclear. Furthermore, ELK1 has been suggested to have a carcinogenic role in human cancers, like breast cancer, thyroid cancer, hepatocellular carcinoma, and colorectal cancer.19,22 Emerging evidence has indicated that ELK1 could serve as an oncogene in CC via promoting cancer development.23,24 Here, we found there might be ceRNA cross talk of circ_0000285–miR197-3p–ELK1 because of the potential complementary sequence between them predicted via Circular RNA Interactome and StarBase tools. In this research, we measured circ_0000285 expression in CC and explored the effect of circ_0000285 on CC development in vitro and in vivo. Moreover, we explored the ceRNA network of circ_0000285–miR197-3p–ELK1 in CC cells.
Methods
Patient Tissue
Thirty CC patients were recruited from Zoucheng People’s Hospital. Tumor-tissue and paired adjacent normal-tissue samples were obtained via surgery and stored at −80°C until use. The patients did not receive other therapy before tissue collection. The clinical features of patients are shown in Table 1. All patients provided written informed consent. This study was permitted via the Ethics Committee of Zoucheng People’s Hospital.
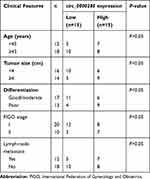 |
Table 1 Associations between circ_0000285 expression and clinical features of CC patients |
Cell Culture
Human CC cell lines (SiHa and HeLa) and normal cervical epithelial cells (H8) were obtained from BeNa Culture Collection (Beijing, China). DMEM (HyClone, Logan, UT, USA) was used for SiHa cells and RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) for HeLa and H8 cells. All cells were maintained at 37°C and 5% CO2 in complete medium with 10% FBS and 1% antibiotic (Thermo Fisher Scientific).
RNA Extraction and Quantitative Reverse-Transcription Polymerase Chain Reaction
RNA was isolated with Trizol (Sigma-Aldrich, St Louis, MO, USA). Nuclear and cytoplasmic RNA was extracted with a Paris kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. For circRNA extraction, RNA was further incubated with RNase R (GeneSeed, Guangzhou, China) at 37°C. RNA was reverse-transcribed to cDNA using a specific reverse-transcription kit (Thermo Fisher Scientific). cDNA was mixed with SYBR Green (Thermo Fisher Scientific) and specific primers, then applied to qRT-PCR on an ABI 7900 system (Foster City, CA, USA). Primers were generated from Sangon (Shanghai, China): circ_0000285 (sense, 5ʹ-TACCTCTGCAGGCAGGAACT-3ʹ; antisense, 5ʹ-TCACATGAATTTAGGTGGGACTT-3ʹ), linear_0000285 (sense, 5ʹ-TGGATATTTGTAAGTCCCACCT-3ʹ; antisense, 5ʹ-TGTGGTCAATGCCTGACTTC-3ʹ), ELK1 (sense, 5ʹ-TCAACTTTCAGGAGACCCGT-3ʹ; antisense, 5ʹ-TGGCATGGTGGAGGTAACAG-3ʹ), miR197-3p (sense, 5ʹ-ACACTCCAGCTGGGTTCACCACCTTCTCCA-3ʹ; antisense, 5ʹ-TCGTGGAGTCGGCAATTCAGTTGAGGCT-3ʹ), U6 (sense, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; antisense, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ), and GAPDH (sense, 5ʹ-CTCTGCTCCTCCTGTTCGAC-3ʹ; antisense, 5ʹ-AAATGAGCCCCAGCCTTCTC-3ʹ). GAPDH (for circ_0000285, linear_0000285, ELK1, or cytoplasm) and U6 (for miR197-3p or nuclear RNA) served as internal controls. Relative RNA expression was determined by the 2–ΔΔCt method.25
Cell Transfection
A circ_0000285-overexpression vector was constructed using a pCD5-ciR vector (GeneSeed), with a pCD5-ciR vector alone as negative control (NC; vector). An ELK1-overexpression vector (pc-ELK1) was constructed using a pcDNA3.1 vector (Thermo Fisher Scientific). The empty pcDNA3.1 vector was exploited as NC (pc-NC). siRNA for circ_0000285 (si-circ_0000285, 5ʹ-CCCCAGCUAUUCAAGUGUAAA-3ʹ), siRNA NC (si-NC; 5ʹ-AAGACAUUGUGUGUCCGCCTT-3ʹ), miR197-3p mimic (5ʹ-UUCACCACCUUCUCCACCCAGC-3ʹ), miRNA NC (miRNA NC, 5ʹ-CGAUCGCAUCAGCAUCGAUUGC-3ʹ), miR197-3p inhibitor (5ʹ-GCUGGGUGGAGAAGGUGGUGAA-3ʹ), and inhibitor NC (5ʹ-CUAACGCAUGCACAGUCGUACG-3ʹ) were synthesized via GenePharma (Shanghai, China). CC cells were transfected with the constructed vectors (600 ng) or oligonucleotides (40 nM) using Lipofectamine 3000 reagent (Thermo Fisher Scientific) for 24 hours.
MTT and Colony-Formation Analysis
Cell viability was assessed via MTT assays. SiHa and HeLa cells (104 cells/well) were added to 96-well plates and incubated for 48 hours. At the end point, MTT (Beyotime, Shanghai, China) at a final concentration of 0.5 mg/mL was injected and cells maintained for 4 hours. Next, the medium was changed to 200 μL dimethyl sulfoxide (Beyotime). Absorbance at 570 nm was examined with a microplate reader (Bio-Gene Technology, Guangzhou, China). For colony-formation assays, SiHa and HeLa cells (500 cells/well) were added to six-well plates and incubated for 10 days. The colonies were mixed with methanol (Sigma-Aldrich) and stained with 1% crystal violet (Solarbio, Beijing, China). Colony-formation numbers were counted under microscopy (Nikon, Tokyo, Japan).
Flow Cytometry
Cell apoptosis and cycle distribution were determined via flow cytometry. An annexin V–FITC apoptosis kit (Solarbio) was used for cell-apoptosis assays. SiHa and HeLa cells (105 cells/well) were added to 12-well plates and cultured for 48 hours. Subsequently, cells were harvested with trypsin and resuspended in the binding buffer, followed by incubation of 10 μL annexin V–FITC and propidium iodide (PI) for 10 minutes. Stained cells were measured with flow cytometry (Agilent, Hangzhou, China), and apoptotic rate was presented as the percentage of cells (Annexin V-FITC+ and PI−/+). For cell-cycle assays, SiHa and HeLa cells (2×105 cells/well) were placed in 12-well plates and cultured for 48 hours. Next, cells were collected, fixed via 70% ethanol (Sigma-Aldrich), and interacted with 50 μg/mL PI and RNase A for 20 minutes. Cell-cycle distribution was detected using flow cytometry.
Western Blot
Protein was isolated with RIPA lysis buffer (Thermo Fisher Scientific) and quantified with a BCA-assay kit (Sigma-Aldrich). Protein samples were subjected to SDS-PAGE and transfer of polyvinylidene fluoride membranes (Solarbio). The membranes were blocked in 5% nonfat milk, interacted with primary and secondary antibodies, and then incubated with ECL Western blotting substrate (Solarbio). GAPDH was regarded as a reference, and relative protein expression was normalized to the control group. Antibodies were provided by Abcam (Cambridge, UK): anti–cyclin D1 (ab226977, 1:5,000 dilution), anti-BCL2 (ab194583, 1:500 dilution), anti-PCNA (ab15497, 1:2,000 dilution), anti-LC3II/I (ab51520, 1:1,000 dilution), anti-ELK1 (ab131465, 1:500 dilution), anti-GAPDH (ab70699, 1:5,000 dilution), and horseradish peroxidase–conjugated IgG (ab6721, 1:10,000 dilution).
Dual Luciferase–Reporter Analysis, RNA Immunoprecipitation, and RNA Pull-Down
The complementary site between circ_0000285 and miR197-3p was predicted using Circular RNA Interactome (https://circinteractome.nia.nih.gov) and that between miR197-3p and ELK1 via starBase (http://starbase.sysu.edu.cn). Wild-type (WT) circ_0000285 and WT ELK1 were constructed by inserting the sequence of circ_0000285 or ELK1 3ʹUTR containing miR197-3p binding sites into psiCheck-2 (Promega, Madison, WI, USA). Mutant (Mut) circ_0000285 and Mut ELK1 were generated via mutating the binding sites to CACCACU or ACCACU, respectively. WT or Mut luciferase-reporter vectors (600 ng) were transfected into SiHa and HeLa cells for luciferase-activity assays with a dual-luciferase assay system (Promega).
For RIP assays, the Magna RIP RNA-binding protein-immunoprecipitation kit (Sigma-Aldrich) was used. SiHa and HeLa cells (107 cells) were lysed in lysis buffer and interacted with magnetic beads coated with anti-Ago2 for 8 hours. Anti-IgG was used as NC. RNA on the beads was purified and abundance of circ_0000285, miR197-3p, and ELK1 detected with qRT-PCR. For RNA pull-down assays, a magnetic RNA-protein pull-down kit (Thermo Fisher Scientific) was used. Biotinylated miR197-3p mimic (biotin-miR197-3p-WT), mutant (biotin-miR197-3p-Mut) and NC (biotin-miRNC) were formed with RiboBio (Guangzhou, China). SiHa and HeLa cells (107 cells) transfected with biotin-miR197-3p-WT, biotin-miR197-3p-Mut, or biotin-miRNC were lysed and incubated with magnetic beads overnight. RNA on the beads was eluted and used for detection of circ_0000285 levels via qRT-PCR.
Xenograft Model
BALB/c nude mice (5-week-old females) were obtained from Beijing Laboratory Animal Center (Beijing, China). The lentiviral vector of short-hairpin RNA for circ_0000285 (sh-circ_0000285) or NC (sh-NC) was synthesized via RiboBio. HeLa cells were transfected with sh-circ_0000285 or sh-NC, and stably transfected cells were selected with puromycin. HeLa cells (5×106 cells/mouse) stably transfected with sh-circ_0000285 or sh-NC were subcutaneously inoculated into mouse flanks (n=5/group). Tumor volume was examined every week for 4 weeks and calculated as volume (mm3) =length (mm) × width2 (mm2)/2. At the end of this experiment, mice were euthanized via cervical dislocation under isoflurane anesthesia. Tumor samples were weighed and then exploited for detection of circ_0000285, miR197-3p, and ELK1 levels. Animal treatment met the standards of the Guide for the Care and Use of Laboratory Animals (People’s Republic of China National Standard GB/T 35,892–2018). Animal experiments were approved via the Animal Care and Use Committee of Zoucheng People’s Hospital.
Statistical Analysis
GraphPad Prism 7 (GraphPad, La Jolla, CA, USA) was exploited for statistical analysis. Three independent experiments were conducted. Data are given as means ± SE. Differences were compared with Student’s t-test or ANOVA, followed via Tukey’s post hoc test as appropriate. Associations between circ_0000285 levels and clinical features of patients were analyzed with χ2-tests. Statistical significance was regarded as P<0.05.
Results
circ_0000285 Expression Elevated in CC
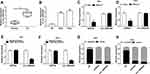
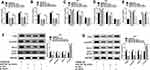
To measure the level of circ_0000285 in CC, we collected 30 paired cancer- and adjacent normal-tissue samples. qRT-PCR data revealed circ_0000285 levels were evidently enhanced in CC tissue when compared to the normal group (Figure 1A). Patients were divided into high and low circ_0000285-expression groups according to the median value of circ_0000285. High expression of circ_0000285 was associated with tumor size, differentiation, and International Federation of Gynecology and Obstetrics stage, but not with age or lymph-node metastasis (Table 1). circ_0000285 levels were higher in CC cell lines (SiHa and HeLa) than normal cervical epithelial cells (H8, Figure 1B). To identify the circular structure of circ_0000285, RNA was treated with RNase R. Results showed that circ_0000285 was more resistant to RNase R than linear GAPDH (Figure 1C and D). Meanwhile, with reverse transfection using random primers or oligo(dT)18 primers, we found that circ_0000285 did not have polyadenylation (Figure 1E and F). In addition, circ_0000285 was mainly located in cytoplasm of SiHa and HeLa cells (Figure 1G and H). These data indicated that high expression of circ_0000285 might have an important role in CC development.
circ_0000285 Knockdown Inhibits Viability, Colony Formation, Arrests Cell Cycle, and Induces Apoptosis and Autophagy in CC Cells
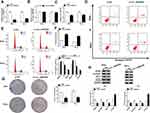
To explore the function of circ_0000285 in CC development, loss-of-function experiments were performed in vitro by knockdown of circ_0000285. Efficacy >60% for circ_0000285 knockdown using si-circ_0000285 was validated in SiHa and HeLa cells (Figure 2A). Meanwhile, si-circ_0000285 did not alter levels of linear_0000285 (Figure 2B). MTT assays showed that circ_0000285 knockdown obviously decreased the viability of SiHa and HeLa cells (Figure 2C). Moreover, analysis of flow cytometry showed that downregulation of circ_0000285 promoted cell apoptosis and arrested the cell cycle at the G0/G1 phase (Figure 2D–F). Additionally, circ_0000285 silence evidently suppressed the colony-formation ability of SiHa and HeLa cells (Figure 2G). Western blot assays showed that interference of circ_0000285 markedly declined protein levels of PCNA, cyclin D1 and BCL2 and enhanced LC3-II/I levels in the two cell lines (Figure 2H). These results suggested that circ_0000285 knockdown repressed CC development in vitro.
circ_0000285 is a Sponge for miR197-3p
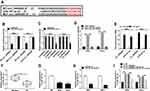
Having got circ_0000285 mainly located in cytoplasm, we wanted to explore whether circ_0000285 could act as an miRNA sponge or ceRNA. Targets of circ_0000285 were predicted via Circular RNA Interactome, which indicated that miR197-3p might be a potential target of circ_0000285 (Figure 3A). To confirm the target association between circ_0000285 and miR197-3p, we constructed WT circ_0000285 and Mut-circ_0000285. Dual luciferase–reporter assays showed that miR197-3p overexpression decreased luciferase activity >50%in the WT circ_0000285 group, but the suppressive effect was abolished in the Mut-circ_0000285 group (Figure 3B). Meanwhile, we explored the impact of miR197-3p on luciferase activity of nontargets (circABCC2, circLRP6, and circSCAF11). Results showed that miR197-3p did not affect the activity of nontargets (Figure 3C). Moreover, RNA pull-down and Ago2 RIP analyses displayed circABCC2 bound with miR197-3p (Figure 3D and E). In addition, lower miR197-3p levels were found in CC tissue and cells than normal tissue and H8 cells, respectively (Figure 3F and G). Furthermore, the influence of circ_0000285 on miR197-3p expression was assessed. The efficacy of miR197-3p inhibitor is validated in Figure 3H. miR197-3p level was elevated via silencing of circ_0000285, which was weakened via knockdown of miR197-3p (Figure 3I). These findings indicated that circ_0000285 was a sponge for miR197-3p in CC cells.
miR197-3p Knockdown Reverses the Effect of circ_0000285 Silence on CC Development in vitro
Next, we explored whether miR197-3p was required for circ_0000285-mediated regulation of CC development. As displayed in Figure 4A and B, miR197-3p knockdown restored silence of circ_0000285-induced inhibition of cell viability and colony-formation ability in SiHa and HeLa cells. Furthermore, downregulation of miR197-3p weakened the knockdown of circ_0000285-induced G0/G1-phase arrest and cell apoptosis (Figure 4C–E). miR197-3p inhibition attenuated the regulatory effect of circ_0000285 interference on protein levels of PCNA, cyclin D1, BCL2, and LC3-II/I in SiHa and HeLa cells (Figure 4F). These data indicated that circ_0000285 knockdown suppressed CC development in vitro by increasing miR197-3p.
ELK1 is a Target of miR197-3p
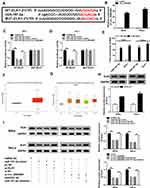
To probe the ceRNA network further, targets of miR197-3p were searched. StarBase predicted ELK1 as a candidate target of miR197-3p. To identify the target association between miR197-3p and ELK1, WT-ELK1 and Mut-ELK1 were constructed (Figure 5A). The influence of circ_0000285 and miR197-3p on luciferase activity was analyzed. Transfection efficacy of the circ_0000285-overexpression vector is confirmed in Figure 5B. miR197-3p upregulation obviously decreased luciferase activity in the WT-ELK1 group, which was restored via upregulation of circ_0000285, while neither circ_0000285 nor miR197-3p affected the activity in the Mut-ELK1 group (Figure 5C and D). In addition, Ago2 RIP assays showed that miR197-3p and ELK1 were enriched in the same complex (Figure 5E). The Cancer Genome Atlas (TCGA) database showed that ELK1 expression is enhanced in CC tissue (Figure 5F and G). Furthermore, the effect of circ_0000285 and miR197-3p on ELK1 expression was investigated. Transfection efficacy of pc-ELK1 is validated in Figure 5H. As shown in Figure 5I, ELK1-protein levels were evidently reduced via miR197-3p overexpression and restored via introduction of pc-ELK1. circ_0000285 knockdown significantly decreased ELK1-protein expression, and this effect was weakened via downregulation of miR197-3p. These data indicate that ELK1, as a target of miR197-3p, was regulated via circ_0000285 through competitively binding with miR197-3p.
miR197-3p Overexpression Represses Viability and Colony Formation, Arrests Cell Cycle, and Facilitates Apoptosis and Autophagy via Targeting ELK1 in CC Cells
To explore how and whether miR197-3p took part in CC development in vitro, SiHa and HeLa cells were transfected with miRNA NC, miR197-3p mimic, miR197-3p mimic + pc-NC, or pc-ELK1. Overexpression of miR197-3p remarkably inhibited cell viability and colony formation (Figure 6A and B), induced cell-cycle arrest at the G0/G1 phase (Figure 6C and D), promoted cell apoptosis (Figure 6E), decreased protein levels of PCNA, cyclin D1, and BCL2, and increased LC3-II/I levels (Figure 6F and G) in SiHa and HeLa cells. These events were alleviated via ELK1 upregulation (Figure 6A–G). These findings suggested that miR197-3p repressed CC development by targeting ELK1 in vitro.
Knockdown of circ_0000285 Reduces Xenograft Tumor Growth In Vivo
To explore the role of circ_0000285 in CC development in vivo, HeLa cells stably transfected with sh-circ_0000285 or sh-NC were used to establish a xenograft model, and were classified as sh-circ_0000285 or sh-NC (n=5). Tumor volume was examined every week and tumor weight measured at the end point. As shown in Figure 7A and B, tumor volume and weight were evidently reduced in the sh-circ_0000285 group in comparison to the sh-NC group. Furthermore, expression levels of circ_0000285, miR197-3p and ELK1 were examined in each group at the end point. As displayed in Figure 7C–F, circ_0000285 and ELK1 levels were markedly decreased and miR197-3p expression enhanced in the sh-circ_0000285 group in comparison to the sh-NC group.
Discussion
CC is a global public health problem in women.26 circRNAs can be used as important biomarkers for the development and treatment of CC.8 In this work, circ_0000285 was upregulated in CC, consistent with a previous study.14 This indicated that high expression of circ_0000285 might be associated with CC malignancy. Our study confirmed that circ_0000285 knockdown suppressed CC development in vitro and in vivo. Moreover, here we are the first to identify the potential ceRNA network of circ_0000285–miR197-3p–ELK1 in CC.
Previous research has suggested that circ_0000285 knockdown repressed CC-cell proliferation via decreasing cell viability and arresting the cell cycle at the G0/G1 phase.14 These results were also confirmed in our study. Moreover, colony formation, apoptosis, and autophagy were also related to cell growth and malignancy in CC.27,28 By performing related experiments and detecting related biomarkers,27,30 we found that circ_0000285 silence inhibited CC-cell growth by reducing colony formation and promoting apoptosis and autophagy. These data implied the carcinogenic role of circ_0000285 in CC, which was also in agreement with that in other cancers.10,12,13 Nevertheless, the mechanism addressed via circ_0000285 in CC development needs more exploration.
The ceRNA network is an important mechanism underlying the role of circRNA located at cytoplasm in cancers.31 A study has indicated circ_0000285 can function as a ceRNA for miR599 to regulate TGFβ2.12 Our study is the first to confirm circ_0000285 is a sponge for miR197-3p via dual luciferase–reporter assays, RIP, and RNA pull-down. In this research, downregulated miR197-3p was measured in CC, consistent with former work.18 That paper showed that miR197-3p had a tumor-suppressive role in CC by suppressing proliferation and invasion.18 Similarly, our research also identified the antitumor effect of miR197-3p in CC via regulating cell viability, colony formation, the cell cycle, apoptosis, and autophagy. However, this function iss opposite to that in some cancers, such as thyroid cancer and bladder cancer.16,32 We hypothesized this might result from altered tumor microenvironments in different cancers. Furthermore, the rescue experiments indicated that circ_0000285 regulated CC development via sponging miR197-3p.
To analyze ceRNA cross talk further, we explored and analyzed targets of miR197-3p. Previous studies have validated multiple targets of miR197-3p in different cancers, such as FoxM1, KLF10, and PKCβ.18,32,33 In this research, we were the first to identify miR197-3p directly targeting ELK1, an oncogene in CC predicted via the TCGA. Former work has indicated that ELK1 can promote CC development via inducing proliferation, migration, and invasion and inhibiting apoptosis and autophagy.24,34 Similarly, this study also uncovered the oncogenic role of ELK1 in CC cells by reversing the anticancer role of miR197-3p. This also indicated that miR197-3p regulated CC development via directly targeting ELK1. In addition, through dual luciferase–reporter assays and Western blot, we found that circ_0000285 modulated ELK1 expression by competitively binding with miR197-3p, implying the ceRNA network of circ_0000285–miR197-3p–ELK1 in CC in vitro. Also, we identified the anticancer role of circ_0000285 knockdown in CC in vivo using a murine xenograft model, also in agreement with a previous study.14 miR197-3p and ELK1 were dysregulated in xenograft tumor tissue, indicating that miR197-3p and ELK1 might also explain circ_0000285 function in vivo.
In conclusion, circ_0000285 silence inhibited CC development, possibly via regulating miR197-3p and ELK1 in a ceRNA-based mechanism. This study indicates a new mechanism for understanding the pathogenesis of CC and suggests that circ_0000285 might be a target for the treatment of CC.
Disclosure
The authors report no funding and no conflicts of interest in this work.
References
1. Cohen PA, Jhingran A, Oaknin A, et al. Cervical cancer. Lancet. 2019;393(10167):169–182. doi:10.1016/S0140-6736(18)32470-X
2. Torre LA, Siegel RL, Ward EM, et al. Global cancer incidence and mortality rates and trends--an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27. doi:10.1158/1055-9965.EPI-15-0578
3. Johnson CA, James D, Marzan A, et al. Cervical cancer: an overview of pathophysiology and management. Semin Oncol Nurs. 2019;35(2):166–174. doi:10.1016/j.soncn.2019.02.003
4. Huang J, Zhou Q, Li Y. Circular RNAs in gynecological disease: promising biomarkers and diagnostic targets. Biosci Rep. 2019;39(5):BSR20181641. doi:10.1042/BSR20181641
5. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
6. Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005. doi:10.1080/15476286.2018.1486659
7. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
8. Chaichian S, Shafabakhsh R, Mirhashemi SM, et al. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2020;235(2):718–724. doi:10.1002/jcp.29009
9. Varghese VK, Shukla V, Kabekkodu SP, et al. DNA methylation regulated microRNAs in human cervical cancer. Mol Carcinog. 2018;16(5):370–382. doi:10.1002/mc.22761
10. Shuai M, Hong J, Huang D, et al. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett. 2018;16(5):6495–6501. doi:10.3892/ol.2018.9471
11. Chi BJ, Zhao DM, Liu L, et al. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma. 2019;66(2):197–202. doi:10.4149/neo_2018_180318N185
12. Zhang Z, Pu F, Wang B, et al. Hsa_circ_0000285 functions as a competitive endogenous RNA to promote osteosarcoma progression by sponging hsa-miRNA-599. Gene Ther. 2019:1–10.
13. Qin J-B, Chang W, Yuan G-H, et al. Circular RNA hsa_circ_0000285 acts as an oncogene in laryngocarcinoma by inducing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(24):10803–10809. doi:10.26355/eurrev_201912_19783
14. Chen RX, Liu HL, Yang LL, et al. Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci. 2019;23(20):8771–8778. doi:10.26355/eurrev_201910_19271
15. Nahand JS, Taghizadeh-Boroujeni S, Karimzadeh M, et al. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol. 2019;234(10):17064–17099. doi:10.1002/jcp.28457
16. Liu K, Huang W, Yan D-Q, et al. Overexpression of long intergenic noncoding RNA LINC00312 inhibits the invasion and migration of thyroid cancer cells by down-regulating microRNA-197-3p. Biosci Rep. 2017;37(4):BSR20170109. doi:10.1042/BSR20170109
17. Ni JS, Zheng H, Huang ZP, et al. MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol Lett. 2019;17(2):2317–2327. doi:10.3892/ol.2018.9848
18. Hu Q, Du K, Mao X, et al. miR-197 is downregulated in cervical carcinogenesis and suppresses cell proliferation and invasion through targeting forkhead box M1. Oncol Lett. 2018;15(6):10063–10069. doi:10.3892/ol.2018.8565
19. Ahmad A, Zhang W, Wu M, et al. Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Genes Genomics. 2018;40(3):243–251. doi:10.1007/s13258-017-0624-6
20. Kong Y, Yin J, Fu Y, et al. Suppression of Elk1 inhibits thyroid cancer progression by mediating PTEN expression. Oncol Rep. 2018;40(3):1769–1776. doi:10.3892/or.2018.6554
21. Fan H-X, Feng Y-J, Zhao X-P, et al. MiR-185-5p suppresses HBV gene expression by targeting ELK1 in hepatoma carcinoma cells. Life Sci. 2018;213:9–17. doi:10.1016/j.lfs.2018.10.016
22. Fan C, Lin B, Huang Z, et al. MicroRNA-873 inhibits colorectal cancer metastasis by targeting ELK1 and STRN4. Oncotarget. 2019;10(41):4192–4204. doi:10.18632/oncotarget.24115
23. Zhao H, Hu G-M, Wang W-L, et al. LncRNA TDRG1 functions as an oncogene in cervical cancer through sponging miR-330-5p to modulate ELK1 expression. Eur Rev Med Pharmacol Sci. 2019;23(17):7295–7306. doi:10.26355/eurrev_201909_18834
24. Tang Q, Chen Z, Zhao L. Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging. 2019;11(22):9982–9999. doi:10.18632/aging.102356
25. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
26. Small W
27. Gao J, Yu H, Guo W, et al. The anticancer effects of ferulic acid is associated with induction of cell cycle arrest and autophagy in cervical cancer cells. Cancer Cell Int. 2018;18(1):102. doi:10.1186/s12935-018-0595-y
28. Huang S, Xie T, Liu W. Icariin inhibits the growth of human cervical cancer cells by inducing apoptosis and autophagy by targeting mTOR/PI3K/AKT signalling pathway. J BUON. 2019;24(3):990–996.
29. Wang S-C. PCNA: a silent housekeeper or a potential therapeutic target? Trends Pharmacol Sci. 2014;35(4):178–186. doi:10.1016/j.tips.2014.02.004
30. Lu H-J, Jin P-Y, Tang Y, et al. microRNA-136 inhibits proliferation and promotes apoptosis and radiosensitivity of cervical carcinoma through the NF-κB pathway by targeting E2F1. Life Sci. 2018;199:167–178. doi:10.1016/j.lfs.2018.02.016
31. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
32. Li Z, Hong S, Liu Z. LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem Biophys Res Commun. 2018;503(3):1825–1829. doi:10.1016/j.bbrc.2018.07.120
33. Chen Z, Ju H, Zhao T, et al. hsa_circ_0092306 targeting miR-197-3p promotes gastric cancer development by regulating PRKCB in MKN-45 cells. Mol Ther Nucleic Acids. 2019;18:617–626. doi:10.1016/j.omtn.2019.08.012
34. Zhang P, Kong F, Deng X, et al. MicroRNA-326 suppresses the proliferation, migration and invasion of cervical cancer cells by targeting ELK1. Oncol Lett. 2017;13(5):2949–2956. doi:10.3892/ol.2017.5852
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.