Back to Journals » OncoTargets and Therapy » Volume 11
Downregulation and DNA methylation of ECRG4 in gastric cancer
Authors Deng P, Chang XJ , Gao ZM, Xu XY, Sun AQ, Li K, Dai DQ
Received 31 December 2017
Accepted for publication 26 March 2018
Published 13 July 2018 Volume 2018:11 Pages 4019—4028
DOI https://doi.org/10.2147/OTT.S161200
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Peng Deng,1 Xiao-Jing Chang,1 Zi-Ming Gao,2 Xiao-Yang Xu,1 An-Qi Sun,2 Kai Li,2 Dong-Qiu Dai1
1Department of Gastrointestinal Surgery and Cancer Center, The Fourth Affiliated Hospital of China Medical University, Shenyang, China; 2Department of Surgical Oncology and General Surgery, The First Affiliated Hospital of China Medical University, Shenyang, China
Background: Esophageal cancer-related gene 4 (ECRG4) is a novel candidate tumor suppressor gene. Our study investigated the expression and function of ECRG4 in gastric cancer and highlighted the role of DNA hypermethylation at the promoter in silencing the ECRG4 expression.
Methods: The GSE63089 data set was obtained from the Gene Expression Omnibus and analyzed for differentially expressed genes. Carcinoma and para-carcinoma tissues of 102 patients with gastric cancer were collected from January 2010 to July 2011. Immunohistochemistry, real-time polymerase chain reaction (PCR), and western blot analyses were performed to evaluate the expression of ECRG4. After measuring the change in the level of ECRG4 expression, CCK-8, Transwell, and flow cytometric cell cycle assays were performed. In addition, methylation-specific PCR was performed to detect the methylation state of ECRG4, and 5-aza-2'-deoxycytidine was used for demethylation of ECRG4. All statistical analyses were performed using the SPSS 17.0 software.
Results: We found that ECRG4 expression was downregulated in gastric cancer, and this was closely related to lymph node metastasis. After ECRG4 was silenced using a specific small interfering RNA, the BGC-823 cell line became highly aggressive and proliferative. In addition, we verified whether downregulation of ECRG4 was highly correlated with DNA methylation of the ECRG4 promoter and found that the demethylating agent 5-aza-2'-deoxycytidine could effectively enhance ECRG4 expression.
Conclusion: The aberrant expression of ECRG4 is associated with hypermethylation in the promoter region and plays an important role in the malignancy of gastric cancer. Therefore, ECRG4 may be a potential biomarker for molecular diagnosis of gastric cancer, and the use of 5-Aza-dC to reverse the hypermethylation of ECRG4 may be a new approach to the treatment of gastric cancer.
Keywords: gastric cancer, ECRG4, DNA methylation, 5-Aza-dC
Introduction
Gastric cancer (GC) is a major health problem worldwide, as it is the fourth most common malignancy and the second cause of death among carcinomas.1,2 Besides genetic alterations, the occurrence of GC is also associated with epigenetic mechanisms such as DNA methylation and histone modifications. DNA methylation of cytosine residues in CpG islands can occur in the promoter regions of genes, regulating cell proliferation, apoptosis, and DNA repair. Studies have reported that DNA methylation is a major mechanism for the inactivation of tumor suppressor genes (TSGs), and it is therefore a target for the detection and diagnosis of cancers.3–5
Esophageal cancer-related gene 4 (ECRG4), also known as chromosome 2 open reading frame 40 (c2orf40), is a novel TSG, which was first found to be downregulated in esophageal cancer. Studies have demonstrated that the ECRG4 protein is downregulated in several tumors, such as breast cancer, colorectal cancer, and glioma,6–11 and the expression level of ECRG4 may be associated with regional lymph node metastasis, tumor stage, and even survival time.12–14 However, the role of ECRG4 in GC is debatable.
In this study, we have highlighted the role of ECRG4 expression and its DNA methylation level in GC tumorigenesis and have provided evidence that ECRG4 might be a new biomarker and treatment target for GC in the future.
Materials and methods
Microarray data
Public data sets of gene expression profiles were obtained from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). The GSE63089 data set was identified, which includes the expression data for 45 paired GC tissues and gastric normal tissues.15 Differential analysis was performed by using GEO2R, and partial results were presented as a heatmap by using the Morpheus tool (https://software.broadinstitute.org/morpheus). We screened the data for differentially expressed genes, with p<0.01, adjusted p<0.01, and |log FC| >1, between the GC and normal tissues.
Cell lines
The human GC cell lines BGC-823, SGC-7901, MKN-45, and AGS and the immortalized normal gastric cell line (GES1) were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China).
Tissue specimens and clinicopathological characteristics
GC carcinoma and paracarcinoma tissues were collected from 102 patients after surgical resection at the First Affiliated Hospital of China Medical University from January 2010 to July 2011. This research was approved by the Ethics Committee of China Medical University, and written informed consent for this study was obtained from each patient. No neoadjuvant radiotherapy, chemotherapy, or targeted therapy was applied. The clinical characteristics included age, gender, tumor size, differentiation state, depth of invasion, tumor location, Borrmann type, and lymph node metastasis.
Reagents
A real-time PCR reverse transcriptase kit was obtained from Fermentas (Vilnius, Lithuania). ECRG4 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) real-time PCR primers and ECRG4 methylation and demethylation primers were synthesized by BGI Company (Shenzhen, China). A rabbit anti-human ECRG4 monoclonal antibody (RabMAb) and ECRG4 isotope antibody were acquired from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). A ready-to-use immunohistochemistry (IHC) hypersensitive Ultra-Sensitive™ SP kit (KIT-9701) and 3,3′-diaminobenzidine kit were purchased from MXB biotechnologles (MXB, Fuzhou, China). A bicinchoninic acid (BCA) protein assay kit was provided by Beyotime (Shanghai, China), and Cell Counting Kit-8 (CCK-8) was from Fanbo Biochemicals Co. Ltd. (Beijing, China). A Wizard DNA cleanup system kit was purchased from Promega Corporation (Fitchburg, WI, USA). Ribonuclease was purchased from Hoffman-La Roche Ltd. (Basel, Switzerland) and nylon mesh from BD Biosciences (San Jose, CA, USA).
IHC
IHC staining of ECRG4 in carcinoma and paracarcinoma gastric tissues was performed by using ECRG4 RabMAb. Global ECRG4 was scored using a semiquantitative integration method as follows: 1) the percentage of positive cells: 0 (≤5%), 1 (6%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (>75%); 2) the intensity of staining: 0 (no staining), 1 (faint yellow), 2 (yellow), and 3 (brown). After multiplying the scores for percentage of positive cells and intensity of staining, the combined staining score was determined as follows: 0 (negative, −), 1–4 (weakly positive, 1+), 5–8 (positive, 2+), and 9–12 (strongly positive, 3+). The data were used for statistical analysis.
Western blotting (WB)
Cells were lysed in a protein lysis buffer, and total protein concentrations were determined by using the BCA assay. Protein lysates were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and proteins were electrotransferred onto membranes. The membranes were probed with the RabMAb (1:1,000) as a primary antibody, and the mouse anti-human GAPDH primary antibody (1:1,000) was used as a loading control. After washing three times with Tris-buffered saline containing Tween 20, a corresponding secondary antibody was used. Each experiment was performed in triplicate.
Quantitative real-time polymerase chain reaction (qRT-PCR)
For qRT-PCR, total RNA was isolated from cells and tissues by using the TRIzol reagent. cDNA was synthesized from the RNA templates by using the M-MuLV reverse transcriptase and oligo (dT)18 primers. The sense and antisense primers for ECRG4 were 5′-GGTACCAGCAGTTTCTCTACATG-3′ and 5′-CAGCGTGTGGCAAGTCATGGTTAGT-3′, respectively. GAPDH was used as an internal control, and its sense and antisense primers were 5′-AGTCCTTCCACGATACCAAAGT-3′ and 5′-AGTCCTTCCACGATACCAAAGT-3′, respectively. The PCR mixture (25 μL) was pre-denatured at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. Each experiment was repeated three times. Analysis was performed by using the 2−ΔΔCt relative quantitation method.
Transfection
An ECRG4-specific siRNA and pcDNA3.1-ECRG4 vector were transfected into BGC-823 cells to silence and enhance the expression of ECRG4, respectively. Groups transfected with an empty vector and without a vector were used as controls. Transfection was performed by using the Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA). The expression level of ECRG4 was confirmed by WB.
Cell proliferation analysis
Cell proliferation was determined by using the CCK-8 assay. A cell suspension (5×105) was seeded into 96-well plates, and the plates were incubated at 37°C, 5% CO2 overnight. The CCK-8 reagent (10 μL) was added to each well, and the plates were incubated for another 3 h. Cell viability was determined by measuring absorbance values at 450 nm.
Transwell assay
A total of 1×104 cells were added to 200 μL of RPMI-1640 medium with 1% newborn calf serum (NBCS), and the suspension was seeded in the upper chamber of a Transwell device. To the lower chamber, 500 μL of RPMI-1640 medium with 20% NBCS was added as a chemoattractant. After incubation for 24 h, the cells on the top surface of the insert were removed by using a cotton swab. Then, the insert was placed in a stationary liquid for 30 min, stained with Giemsa stain for 8 min, washed, and finally observed under a microscope.
Flow cytometric cell cycle analysis
Cells were trypsinized, collected, and washed with cold PBS before being fixed with 70% ethanol at −20°C overnight. The fixed BGC-823 cells were washed in cold PBS, and then incubated at 37°C with 100 ng/mL ribonuclease and 10 ng/mL propidium iodide (PI) for 30 min. After filtering through a nylon mesh, samples were analyzed with a FACScan flow cytometer, and data were analyzed with ModFit software (Verity Software House, Topsham, ME, USA).
Methylation-specific PCR (MSP)
We detected the methylation state of ECRG4 by MSP. DNA was isolated from GC cell lines and tissues by using phenol–chloroform–isoamyl alcohol. Then, sodium hydrogen sulfite was removed from DNA by using the Wizard DNA cleanup system kit. The purified DNA was immediately used to perform MSP. The primers for methylated sequences were 5′-TGGCGTTTTTATGGTGTTC-3′ (upstream) and 5′-CACCACTTCGCACTTATACG-3′ (downstream), generating a 137-bp PCR product. The primers for unmethylated sequences were 5′-ATGTGGTGTTTTTATGGTGTTT-3′ (upstream) and 5′-AAACACCACTTCACACTTATACA-3′ (downstream), generating another 137-bp PCR product. The total reaction volume was 50 μL, and the mixture contained DNA, sense and antisense primers, 10× Dream Taq buffer, dNTP mix, and Dream Taq DNA polymerase. The reaction conditions were as follows: pre-denaturation at 95°C for 5 min, followed by 34 cycles at 95°C for 30 s, 62°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min. The PCR products were subjected to agarose gel electrophoresis at 120 V for 40 min. The gels were photographed by using an electrophoretic gel imaging analysis system.
Cell treatment with 5-Aza-2′-deoxycytidine (5-Aza-dC)
We first maintained BGC-823 cells under starvation conditions in RPMI-1640 medium containing 1% fetal bovine serum and then treated the cells with 1 μmol/L, 5 μmol/L, or 10 μmol/L 5-Aza-dC. The cells were transferred daily into a fresh culture medium containing 5-Aza-dC at the three experimental concentrations. On the third day, the cells were harvested for further analysis. Simultaneously, BGC-823 cells were cultivated without drug treatment as a control.
Statistical analysis
Continuous variables are presented as the mean values and standard deviation. Differences between groups were determined by using an independent sample t-test and χ2 test. Each assay was repeated three times, and p<0.05 was considered statistically significant. All analyses were performed by using the SPSS 17.0 software (SPSS Inc., Chicago, IL, USA).
Results
Expression of ECRG4 is downregulated and closely related to lymph node metastasis in GC
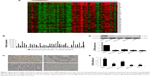
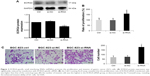
A heatmap of the 100 most downregulated genes in the GSE63089 public data set, which includes expression data from 45 paired GC and normal tissues, is presented in Figure 1A. The expression level of ECRG4 in normal gastric tissues was about 2.46-fold higher than that in GC tissues (log fold change [log FC] = 2.46, p<0.01). A qRT-PCR assay on tissues from 35 patients showed that ECRG4 mRNA was downregulated in GC tissues (p<0.05; Figure 1B). Using an IHC assay, we also observed a lower expression intensity of the ECRG4 protein in GC tissues from 102 patients (Figure 1C). Among the 102 GC patients, 35.3% (38/102) showed a lower expression of ECRG4 in GC tissues, while 3.9% (4/102) showed a lower expression of ECRG4 in normal tissues (p<0.01). The ECRG4 protein was also related to the differentiation state of GC. As shown in Figure 1C, lower ECRG4 expression was always associated with poorer differentiation. To further confirm the ECRG4 expression in cells, we performed WB and qRT-PCR using four GC cell lines. Both the assays demonstrated that protein and mRNA expression of ECRG4 was downregulated in all GC cell lines compared to that in a normal cell line (Figure 1D and E).
Based on the levels of ECRG4 protein and mRNA expression, the 102 GC patients were divided into high and low expression groups. The t-test showed that the ECRG4 protein level in GC was associated with lymph node metastasis (p<0.05) and histological grade (p<0.05) (Table 1). Similarly, the ECRG4 mRNA level in GC was also related to lymph node metastasis (p<0.05), as shown in Table 2. Thus, the expression level of ECRG4 is positively related to lymph node metastasis.
  | Table 1 ECRG4 protein levels and clinicopathological parameters of GC tissues |
  | Table 2 ECRG4 mRNA levels and clinicopathological parameters of GC tissues |
Inhibition of ECRG4 promotes proliferation and invasion of GC cells
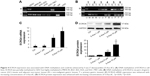
An ECRG4-specific siRNA was transfected into BGC-823 cells, which had a relatively high level of ECRG4 expression. The expression level of ECRG4 in the siRNA-transfected ECRG4 group was significantly reduced compared to that in the non-transfected group or in the group transfected with a negative plasmid (Figure 2A). The CCK-8 assay showed that the proliferation rate of the siRNA-ECRG4 group was higher than that of the other groups (Figure 2B). In addition, the migration ability of BGC-823 cells transfected with siRNA-ECRG4 significantly increased in the Transwell assay, as shown in Figure 2C. These results indicated that the inhibition of ECRG4 expression could enhance the growth and invasion of GC cells.
Effect of ECRG4 expression on the cell cycle of GC cells
To determine the effect of ECRG4 expression in altering the cell cycle in GC cells, BGC-823 cells were transfected with pcDNA3.1, pcDNA3.1-ECRG4, siRNA-control, or siRNA-ECRG4. The cell cycle was analyzed by flow cytometry following PI staining. Compared with the control groups, the percentage of G1 phase cells was significantly increased in the pcDNA3.1-ECRG4 group (70.72% vs 61.95%) and decreased in the siRNA-ECRG4 group (50.89% vs 61.95%) (Figure 3). Our results revealed that ECRG4 expression was associated with variation in the cell cycle in GC cells. In addition, overexpression of ECRG4 induced cycle arrest in the G1 phase and inhibited the proliferation of GC cells, consistent with the results of the CCK-8 assay.
The ECRG4 promoter is highly methylated in GC
We performed the MSP analysis to investigate the methylation status of ECRG4 in GC cells. As shown in Figure 4A, the ECRG4 promoter was highly methylated in four GC cell lines but partially methylated in GES1. In addition, we analyzed the ECRG4 methylation status in 74 GC tissues (Figure 4B) and confirmed that methylation of the ECRG4 promoter was also higher in GC tissues than that in non-tumor tissues (p<0.05). ECRG4 was hypermethylated in 66.2% (49/74) of the carcinoma tissues but only in 18.9% (14/74) of the paracarcinoma tissues. However, there was no correlation between the methylation level of the ECRG4 promoter and clinicopathological characteristics (p>0.05; Table 3).
  | Table 3 Correlation between DNA methylation of ECRG4 and clinicopathological data of GC patients |
The demethylation agent 5-Aza-dC could upregulate ECRG4 expression in GC
5-Aza-dC is an effective suppressor of DNA methylation. After treatment with 5-Aza-dC for 72 h, the expression of ECRG4 mRNA and protein was markedly upregulated in BGC-823 cells (p<0.05; Figure 4C and D). Furthermore, the elevation in ECRG4 expression was positively associated with the drug concentration of 5-Aza-dC. These results indicated that 5-Aza-dC increases the expression of ECRG4 by suppressing its methylation.
Discussion
ECRG4 is a highly conserved gene that plays an important role in tumorigenesis in vertebrates. Studies have revealed the potential TSG role of ECRG4 in different cancers. In our study, we confirmed the association between GC tumorigenesis and ECRG4 expression, particularly, the role of ECRG4 methylation level.
High-throughput analysis showed that ECRG4 might act as a TSG of GC. Using IHC, WB, and qRT-PCR assays, we showed that the expression of ECRG4 mRNA and protein was downregulated in GC, consistent with a tumor-suppressing role of ECRG4. The relationship between clinicopathological characteristics and ECRG4 expression has been reported in several cancers. Studies have revealed that ECRG4 expression is associated with lymph node metastasis, predicting a poor prognosis in patients with tumors. In our study, we also demonstrated the relationship between ECRG4 expression and lymph node metastasis in GC patients. However, a long-term and complete follow-up is needed to confirm the role of ECRG4 as a marker for GC prognosis.
Furthermore, we investigated the tumor-suppressing function of ECRG4 at the cellular level. As previously reported, ECRG4 may be associated with the inhibition of some malignant phenotypes of tumor cells. In our study, we showed that downregulation of ECRG4 promoted proliferation and invasion of GC cells, as demonstrated by CCK-8 and Transwell assays. Notably, Wen et al have found that ECRG4 expression in esophageal cancer is closely associated with the invasive depth and tumor node metastasis stage, also confirming the inhibitory effect of ECRG4 on malignant phenotypes.13 However, the mechanism of effects of ECRG4 effects in tumorigenesis is unclear. Lu et al have reported that ECRG4 may suppress the proliferation and invasion of breast cancer cells by regulating M-phase cell cycle genes.16 The results of our analysis showed that gain or loss of ECRG4 function led to changes in cell cycle in GC cells. Overexpression of ECRG4 induced cell cycle arrest in G1 phase and then suppressed the proliferation of GC cells, which is consistent with previous studies.17,18 In addition, Jia et al have revealed that ECRG4 is associated not only with proliferation but also with apoptosis.17 In laryngeal cancer, overexpression of ECRG4 induced apoptosis by regulating the expression of apoptosis-related factors, such as Bax, Bcl-2, and caspase-3. ECRG4 could also play a role in anti-inflammatory activities by interacting with an immunity receptor complex, which might be important for tumor immunity.19
Promoter DNA methylation plays an important role in the tumorigenesis of various human cancers via long-term silencing of TSGs.5 In esophageal, colorectal, and renal cancers, promoter methylation is the main mechanism of ECRG4 silencing, and treatment with demethylating agents can restore gene expression.20–22 In GC, Huang et al reported that DNA methylation is common in gastric intestinal metaplasia (IM), and hypermethylated regions of IM were recapitulated in GC.23 Moreover, Qu et al reported that methylation levels of ECRG4 were significantly higher in GC tissue (69.4%) than in normal tissue (6.7%).4 In the present study, we confirmed that the ECRG4 promoter was highly methylated, which could explain the downregulation of ECRG4 in GC patients. Although we did not find an association between ECRG4 methylation and clinicopathological characteristics of GC patients, Wang et al22 have reported that aberrant ECRG4 promoter methylation could be a predictor of GC pathological stage. In their study, methylation levels of ECRG4 were higher in stage III/IV (24/30) than in stage I/II (10/19) patients.22
In our study, we found that the demethylating agent 5-Aza-dC enhanced ECRG4 expression by effectively suppressing DNA methylation, and this suppressive effect was positively associated with the drug concentration of 5-Aza-dC. Therefore, reversing the hypermethylation of ECRG4 with 5-Aza-dC might be a new approach for GC treatment. Based on a literature review, 5-Aza-dC, combined with adjuvant chemotherapy, might be an effective measure against GC. For example, Jiang et al have reported that overexpression of ECRG4 may improve the chemosensitivity of the GC-derived SGC-7901 cell line to 5-fluorouracil (5-FU).24 Therefore, we hypothesized that 5-Aza-dC may improve the curative effect of 5-FU to some extent, which would have important clinical significance.
Conclusion
Our results suggested that ECRG4 is a novel TSG for GC, and DNA methylation of ECRG4 is a potential inducer of tumorigenesis. Inhibition of ECRG4 expression may promote the growth and migration of GC cells. Conversely, demethylation of ECRG4 with 5-Aza-dC could suppress the tumorigenesis of GC cells. Thus, restoring the expression of the ECRG4 in the tumor, either by epigenetic therapy or by using a recombinant protein, may offer a promising novel therapeutic approach for GC treatment.
Acknowledgments
This study was funded by the Liaoning Province Science and Technology Plan Project (No. 2013225021) and the Natural Science Foundation of Liaoning Province (No. 201602817). The funders had no role in study design, data collection and analysis, preparation, or publication of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. | ||
Wajed SA, Laird PW, DeMeester TR. DNA methylation: an alternative pathway to cancer. Ann Surg. 2001;234(1):10–20. | ||
Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. | ||
Delpu Y, Cordelier P, Cho W, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14(7):15029–15058. | ||
Cai Z, Liang P, Xuan J, Wan J, Guo H. ECRG4 as a novel tumor suppressor gene inhibits colorectal cancer cell growth in vitro and in vivo. Tumour Biol. 2016;37(7):9111–9120. | ||
Gotze S, Feldhaus V, Traska T, et al. ECRG4 is a candidate tumor suppressor gene frequently hypermethylated in colorectal carcinoma and glioma. BMC Cancer. 2009;9:447. | ||
Li LW, Yu XY, Yang Y, Zhang CP, Guo LP, Lu SH. Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int J Cancer. 2009;125(7):1505–1513. | ||
Li W, Liu X, Zhang B, et al. Overexpression of candidate tumor suppressor ECRG4 inhibits glioma proliferation and invasion. J Exp Clin Cancer Res. 2010;29(1):89. | ||
Matsuzaki J, Torigoe T, Hirohashi Y, et al. Expression of ECRG4 is associated with lower proliferative potential of esophageal cancer cells. Pathol Int. 2013;63(8):391–397. | ||
Sabatier R, Finetti P, Adelaide J, et al. Down-regulation of ECRG4, a candidate tumor suppressor gene, in human breast cancer. PLoS One. 2011;6(11):e27656. | ||
Chen JY, Wu X, Hong CQ, et al. Downregulated ECRG4 is correlated with lymph node metastasis and predicts poor outcome for nasopharyngeal carcinoma patients. Clin Transl Oncol. 2017;19(1):84–90. | ||
Wen Y, Hu X. Expression of esophageal carcinoma related gene 4 (ECRG4) and its clinical significance in prognosis of esophageal carcinoma. Int J Clin Exp Pathol. 2015;8(11):14772–14778. | ||
You Y, Li H, Qin X, Ran Y, Wang F. Down-regulated ECRG4 expression in breast cancer and its correlation with tumor progression and poor prognosis – a short report. Cell Oncol (Dordr). 2016;39(1):89–95. | ||
Zhang X, Ni Z, Duan Z, et al. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS One. 2015;10(2):e0116979. | ||
Lu J, Wen M, Huang Y, et al. C2ORF40 suppresses breast cancer cell proliferation and invasion through modulating expression of M phase cell cycle genes. Epigenetics. 2013;8(6):571–583. | ||
Jia J, Dai S, Sun X, et al. A preliminary study of the effect of ECRG4 overexpression on the proliferation and apoptosis of human laryngeal cancer cells and the underlying mechanisms. Mol Med Rep. 2015;12(4):5058–5064. | ||
Li LW, Li YY, Li XY, Zhang CP, Zhou Y, Lu SH. A novel tumor suppressor gene ECRG4 interacts directly with TMPRSS11A (ECRG1) to inhibit cancer cell growth in esophageal carcinoma. BMC Cancer. 2011;11:52. | ||
Podvin S, Dang X, Meads M, et al. Esophageal cancer-related gene-4 (ECRG4) interactions with the innate immunity receptor complex. Inflamm Res. 2015;64(2):107–118. | ||
Fang WJ, Zheng Y, Wu LM, et al. Genome-wide analysis of aberrant DNA methylation for identification of potential biomarkers in colorectal cancer patients. Asian Pac J Cancer Prev. 2012;13(5):1917–1921. | ||
Luo L, Wu J, Xie J, et al. Downregulated ECRG4 is associated with poor prognosis in renal cell cancer and is regulated by promoter DNA methylation. Tumour Biol. 2016;37(1):1121–1129. | ||
Wang YB, Ba CF. Promoter methylation of esophageal cancer-related gene 4 in gastric cancer tissue and its clinical significance. Hepatogastroenterology. 2012;59(118):1696–1698. | ||
Huang K, Ramnarayanan K, Zhu F, et al. Genomic and epigenomic profiling of high-risk intestinal metaplasia reveals molecular determinants of progression to gastric cancer. Cancer Cell. 2018;33(1):137.e135–150.e135. | ||
Jiang CP, Wu BH, Wang BQ, et al. Overexpression of ECRG4 enhances chemosensitivity to 5-fluorouracil in the human gastric cancer SGC-7901 cell line. Tumour Biol. 2013;34(4):2269–2273. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.