Back to Journals » OncoTargets and Therapy » Volume 12
Downregulated miR-585-3p promotes cell growth and proliferation in colon cancer by upregulating PSME3
Authors Liu C, Yang J, Wu H, Li J
Received 28 January 2019
Accepted for publication 16 July 2019
Published 14 August 2019 Volume 2019:12 Pages 6525—6534
DOI https://doi.org/10.2147/OTT.S203175
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Chunmei Liu,1,2 Juan Yang,1,2 Han Wu,1,2 Jun Li3
1Department of Pathology, Luohe Central Hospital, Luohe 462000, People’s Republic of China; 2Department of Pathology, The First Affiliated Hospital of Luohe Medical College, Luohe 462000, People’s Republic of China; 3Nursing Department, Xiangya Hospital, Central South University, Changsha 410078, People’s Republic of China
Background: Upregulation of PSME3 and its oncogenic roles have been reported in colon cancer recently. However, the underlying mechanism of PSME3 upregulation remains unknown. Here, we explored the expression of PSME3 and subsequently uncovered its mechanism in colon cancer.
Materials and methods: The expression of PSME3 was analyzed by using online databases, Oncomine and UALCAN. qPCR was carried out to detect the expression of PSME3 in collected colon cancer tissues and cell lines. Moreover, the promoter methylation and the hnRNA level of PSME3 were also analyzed by online database and qPCR, respectively. The candidate miRNAs targeting PSME3 were predicted by Starbase 3.0 and validated by luciferase reporter system. CCK-8, plate colon formation, and Edu incorporation were applied to study the functions of miRNA in colon cancer. The expression of miRNA and its correlation with PSME3 were detected in colon cancer tissues.
Results: Oncomine and UALCAN data indicate PSME3 is obviously upregulated in colon cancer tissue samples which is further confirmed in collected colon cancer tissues and cells by qPCR. No significant difference in methylation status promoter of PSME3 was observed between colon and colon cancer tissues. The hnRNA level of PSME3 was comparable between colon epithelial cell and colon cancer cells. miR-585-3p is predicted to directly target PSME3 and is validated by luciferase reporter assay. Then, miR-585-3p downregulation is confirmed and miR-585-3p restoration can suppress cell growth and proliferation by inhibiting PSME3 in colon cancer indicating by CCK-8, plate colon formation, and Edu incorporation. Moreover, negative correlation in expression between miR-585-3p and PSME3 was observed in our collected tissues samples.
Conclusion: We reveal for the first time that miR-585-3p downregulation accounts for the overexpression of PSME3 in colon cancer. Moreover, miR-585-3p, serving as a tumor suppressor, can inhibit cell growth and proliferation in colon cancer by targeting PSME3.
Keywords: colon, cancer, miR-585-3p, PSME3, proliferation
Introduction
Colon cancer is one of the most common diagnosed cancers without sex preference. Although considerable advances have been achieved in treatment in the past few decades, the overall survival time and 5-year survival rate of colon cancer have not been significantly improved. Colon cancer remains a leading cause of tumor-related morbidity and mortality all over the world.1–4 Therefore, it is urgent and meaningful to further identify critical regulators and explore its mechanisms in colon cancer tumorigenesis which may eventually arise novel treatments to improve patient prognosis.
Proteasome activator subunit 3 (PSME3), also referred to REGγ, Ki antigen, and PA28γ, was first cloned in 1990 from a patient with systemic lupus erythematosus.5 The vital functions of PSME3 in the regulation of mitosis, immune response, and cell destiny determination, both at psychological and pathological status, have been revealed by numerous studies.5 Recently, the functions of PSME3 in tumor development and progression have received much attention. Aberrant overexpression of PSME3 has been revealed in a plethora of cancers including pancreatic cancer,6 thyroid carcinoma,7 breast cancer,8 melanoma,9 oral cancer,10 non-small-cell lung cancer,11 and multiple myeloma.12 Essentially, PSME3 severs as an oncogene, indicating by the existing studies, and promotes tumor apoptosis resistance, growth and metastasis by modulating epithelial-mesenchymal transition, stem characteristics, and glycolysis in Myc,6 Hippo or NF-kB13 associated manner. As for colon cancer, high expression of PSME3 and its oncogenic functions have been revealed by a recent study.13 However, the underlying mechanism for PSME3 upregulation has not been elucidated.
MicroRNAs, a class of small no coding RNAs consisting of 19–23 nucleotides, are epigenetic regulators and play vital roles in modulation of gene expression profile.14 Based on base complementation pairing rule, miRNAs could directly bind target mRNAs and subsequently promote mRNAs degradation or translation inhibition by which decrease the protein level in a post-transcriptional manner.15 Therefore, miRNAs nearly exert essential roles in all biological processes. The vital roles of miRNAs in carcinogenesis have been exclusively revealed by numerous studies.15,16 Abnormal expressions of miRNAs, leading to the associated inverse expression level of target proteins, account for the major underlying causes for cancer development and progression. The vital roles of miRNAs, such as miR-214,17 −483,18 −485,19 −210,20 and −21,21 etc, in carcinogenesis of colon cancer by targeting different targets have been exclusively explored.22 According to the previous studies, miR-7, being downregulated in several cancers, can directly target PSME3 and suppress its oncogenic effects in breast cancer and lung cancer.23 Considering miR-7 is the only reported miRNA for PSME3 up to now, we speculate that there may be other miRNAs for targeting PSME3.
In this study, we uncover that miR-585-3p, being downregulated in colon cells and tissues, is a new negative regulator of PSME3. Re-expression of miR-585-3p can inhibit the proliferation and growth of colon cancer cells by directly targeting PSME3. Moreover, miR-585-3p is negatively correlated with PSME3 mRNA in colon cancer tissues. Taken together, our data indicate that loss of miR-585-3p accounts for the upregulated PSME3, which subsequently exerts pro-proliferation functions in colon cancer.
Methods and materials
Tissue samples
20 paired cases of paraffin-embedded colon cancer samples and para-tumor samples, from July 2017 to June 2018, were collected from the newly diagnosed patients, who only receive surgical resection at that time, at General surgery of Luohe Central hospital. Furthermore, clinical pathological features were also collected. The pathological diagnosis of every patient was independently confirmed by two experienced histopathologists. Informed consent for using tissue samples and clinical records was confirmed and obtained from every patient. The patient consents were written informed consent and this study was approved by the Ethics Committee of Louhe Central Hospital. Moreover, the study was conducted according to the guidelines of the Declaration of Helsinki.
Immunohistochemistry
Immunohistochemistry (IHC) was performed using SP Rabbit HRP IHC Kit (CWBIO, Beijing, China) according to its instruction. Briefly, paraffin-embedded sections were deparaffinized by xylene and rehydrated by graded ethanol. After antigen retrieval and endogenous peroxidase activity blocking, the slides were incubated with rabbit anti-PSME3 polyclonal antibody (dilution:1:50, BBI, Shanghai, China) overnight at 4°C. Then, the slides were washed three times with PBS and subsequently incubated with goat anti-rabbit IgG-biotin for 30 min. After three times PBS washing, the slides were incubated with streptavidin-HRP. Afterward, the slides were incubated for 3min in DAB (diaminobenzidine) solution and rinsed twice with H2O. In the end, with counterstaining with hematoxylin, the slides were visualized under a microscope (Olympus, Tokyo, Japan). The IHC results were analyzed using a semiquantitative score system based on staining intensity and area according to the previous description.3
Cell culture
Colon cancer lines, HCT116, SW480, and SW620 were purchased from ATCC and were maintained in RPMI-1640 (BI, Jerusalem, Israel) with 10% fetal bovine serum (FBS, BI, Jerusalem, Israel). Immortalized colon epithelial cell line NCM460, purchased from ATCC, were cultured with M3 Base medium (INCELL, TX, USA) plus 10% FBS (BI, Jerusalem, Israel) 0.293T cells were purchased from the Cell Bank of the Chinese Academy of Sciences and maintained in DMEM (BI, Jerusalem, Israel) with 10% FBS (BI, Jerusalem, Israel). All the cells were grown in a humidified cell incubator at 37 °C with 5% CO2.
Quantitative reverse transcription polymerase chain reaction (qPCR)
Total cell and paraffin-embedded tissue RNA extraction and mRNA reverse transcription were carried out according to the previous study.24 AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) was applied to analyze the expression of miR-585-3p and PSME3 in colon cancer tissues and cells. Simply, SYBR mix, cDNA, specific primers, and deionized water were mixed following the instruction. Then, the PCR reaction was run on the CFX Connect Real-Time System (Bio-Rad, CA, USA) and the results were analyzed with CFX Manager 2.0 (Bio-Rad, CA, USA). The specific primer sequences are listed as follow: PSME3 forward primer: (5ʹ-TCTGACATGAATCTCCCAGTCC-3ʹ), PSME3 reverse primer: (5ʹ- CTCATCCAACCTTCGCTTCTTAT-3ʹ), PSME3 intron 2 forward primer: (5ʹ- GCACCTAGTGTTGATCCTGGT-3ʹ), PSME3 intron 2 reverse primer: (5ʹ- ATCTGCTGGTTCCACAAAGGA-3ʹ), GAPDH forward primer: (5ʹ- CAGCAAGAGCACAAGAGGAA-3ʹ), GAPDH reverse primer: (5ʹ- ATGGTACATGACAAGGTGCGG-3ʹ). Primers for miR-585-3p and U6 were purchased from RioBio Inc (Guangzhou, China).
Transfection
MiR-585-3p mimic and inhibitor, pENTER-PSME3, and corresponding controls were introduced into colon cancer and colon cells by using LipofectamineTM 2000 according to manufactures’ instruction. The miR-585-3p mimic, inhibitor and controls were obtained from RioBio Inc (Guangzhou, China). pENTER-PSME3 and corresponding vector were purchased from Vigene Bioscience Inc (MD, USA).
Western blot
Western bolt was performed as previously described.24 Briefly, RIPA buffer was used to disrupt the cells and the total proteins were prepared via high-speed centrifugation at 4 °C. Then the denatured proteins were separated on 12% SDS PAGE (30 μg/lane) and transferred to polyvinylidene difluoride membranes (PVDF). After immersing in blocking solution (5% non-fat milk) for 1 hr, the membranes were incubated with the follow primary antibodies at 4°C overnight: PMSE3 (dilution: 1:500, BBI, Shanghai, China), GAPDH (dilution: 1:2000, Abclonal, MA, USA) and α-tubulin (dilution: 1:3000, Abclonal, MA, USA). Next day, the membranes were washing by TBST and subsequently incubated with the corresponding secondary antibodies for 1 hr. With another washing process, the membranes were visualized using chemiluminescent HRP substrate (Millipore, MA, USA) via a FluorChem FC3 system (Proteinsimple, CA, USA).
Luciferase reporter assay
Both the wild 3ʹ-UTR sequence, containing predicated complementary region, and the mutant 3ʹ-UTR sequence, lacking with the mutant complementary region, of PSME3 was cloned into psiCHECK2 luciferase reporter vector (Promega Corporation, WI, USA), respectively. Then, luciferase reporter vectors, miR-585-3p mimics, or mimic controls were transfected into 293T cells using HighGene Transfection reagent (Abclonal, MA, USA). After 48 hrs, the luciferase activity was collected by using a Dual-Luciferase Reporter detection System (Promega Corporation, WI, USA). The relative luciferase activity was indicated by the ratio of firefly luciferase to Renilla luciferase activity.
CCK-8 assay
CCK-8 reagent (Beyotime, Shanghai, China) was used to detect the cell growth and viability according to the previous description. Simply, colon cancer cells were seeded in 96-well plates at the density of 1×103 cells/well and cultured for 7 days. Every 24 hrs, 10 μL CCK-8 was added into the well for 1 hr. Then, the absorbance value at 450nm of treated well was detected by Epoch (Bio-Tek, VT, USA). Eventually, the growth curves were drawn based on the absorbance value. Three paralleled wells were made and the assay was repeated in triplicate.
Plate clone formation assay
Plate clone assay was applied to analyze cell proliferation according to the previous paper.3 Briefly, colon cancer cells were seeded into 6-well plates with proper density (800 cells/well). 8 days later, cells were fixed by methanol and then stained with 0.1% crystal violet. Only clones containing more than 50 cells were counted. The plate clone formation assay was performed for three times in triplicate.
Edu incorporation assay
Edu incorporation assay was carried out as described in previous work.3 Briefly, colon cancer cells were successively incubated with 10mM Edu for 2 hrs, fixed by 4% paraformaldehyde, neutralized by 2mg/ml glycine, and permeated by 0.5% TritonX-100. Then, the cells were stained with Apollo staining reaction solution, permeated by 0.5% TritonX-100 again, and stained nucleus by Hoechst 33342. At last, the positive cells with Edu incorporation were visualized under inverted fluorescence microscope (Leica, Solms, Germany).
Statistical analys
All experiments were independently performed for 3 times. Statistical analysis and charts were conducted using GraphPad prism version 6. unpaired t-test or chi-square test were applied to analyze the statistical difference between two groups. Significant differences were considered when P<0.05.
Results
PSME3 mRNA is upregulated in colon cancer
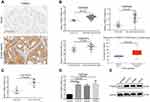
A recent study has demonstrated that PSME3 is upregulated in colon cancer by immunohistochemistry (IHC).13 Indeed, we also observed significant upregulation of PSME3 in colon cancer samples (Figure 1A). Subsequently, we further explored the mRNA level of PSME3 in colon cancer to identify the underlying reason for PSME3 upregulation via online database Oncomine25 and UALCAN.26 As the data from GEO and TCGA shown, significant upregulation of PSME3 is observed in several distinct data sets (Figure 1B). Moreover, upregulated PSME3 is also observed in local colon tissues (Figure 1C). To further validation, we detected the expression of PSME3 in colon cancer cell lines and normal colon epithelial cells. Similarly, colon cancer cell lines, like SW620, HCT-116, and SW480, shown obviously abundant amount of PSME3 mRNA and protein than that in NCM460 (Figure 1D and E). Therefore, these results show that upregulation of PSME3 is due to upregulated mRNA in colon cancer.
miR-585-3p is downregulated and can directly suppress PSME3 in colon cancer cells
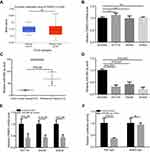
As mentioned above, PSME3 protein upregulation is due to its upregulated mRNA. The mRNA level can be regulated at both transcriptional and post-transcriptional level. Firstly, we analyzed the promoter methylation status, a transcript activity indicator, of PSME3 via UALCAN database,26 and no significant difference was observed between colon tissues and colon cancer tissues (Figure 2A) suggesting that post-transcriptional mechanism may account for upregulated PSME3. Indeed, the level of hnRNA (heterogeneous nuclear RNA) of PSME3, which directly reflect the level of primary PSME3 transcript, were comparable in colon epithelial cell and colon cancer cells (Figure 2B). PSME3 has been reported to be regulated by miR-7 in CHO cells. Therefore, we speculate that miRNAs may exert potential roles in the regulation of PMSE3 mRNA level. Using Starbase 3.0,27 we predicted the potential miRNAs targeting PMSE3. Considering it has the most supporting evidence from 17 sets of Ago CLIP-seq experiments and shows lower expression in colon cancer tissues (Figure 2C), miR-585-3p was selected for further validation. Indeed, downregulated miR-585-3p in colon cancer cells was confirmed by qPCR analysis (Figure 2D). Moreover, miR-585-3p mimics could significantly decrease PSME3 mRNA in colon cancers (Figure 2E). Most importantly, luciferase reporter assay indicated that miR-585-3p could directly target and inhibit PSME3 expression (Figure 2F). Thus, we reveal that miR-585-3p is downregulated and can directly suppress PSME3 in colon cancer.
miR-585-3p mimics inhibit cell growth and proliferation of colon cancer in vitro
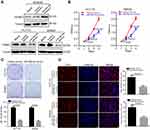
Furthermore, we explored the functions of miR-585-3p in colon cancer. As Figure 3A indicating, miR-585-3p inhibitors significantly increased the level of PSME3 in NCM460, whereas, miR-583-3p mimics successfully decreased the level of PSME3 in colon cancer cells. Then, CCK-8, plate colon formation, and Edu incorporation were carried out to detect the effect of miR-585-3p knockdown on growth and proliferation of colon cancer cells. miR-585-3p mimics significantly suppressed cell growth and proliferation of SW620 and HCT116 demonstrating by lower OD450 value (Figure 3B), fewer colonies (Figure 3C), and decreased Edu incorporation rate (Figure 3D). Thus, these results indicate that miR-585-3p can inhibit the growth and proliferation of colon cancer cells in vitro.
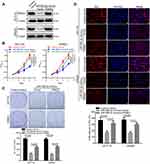
PSME3 restoration antagonizes the inhibitory effects of miR-585-3p on growth and proliferation of colon cancer cells
Furthermore, we explored whether miR-585-3p could exert its effects in colon cancer cells by targeting PSME3. Thus, we explored the influences of ectopic PSME3 expression on the inhibitory effects of miR-585-3p on colon cancers. As Figure 4A indicating, the protein level of PSME3 was successfully restored by ectopic expression. Moreover, ectopic expression of PSME3 can significantly antagonize the inhibitory effects of miR-585-3p on cell growth and proliferation in colon cancer cells (Figure 4B–D). Thus, these results indicate that miR-585-3p can suppress the growth and proliferation by inhibiting PSME3 in colon cancer cells.
miR-585-3p is deregulated and shows a negative correlation to PSME3 in local colon cancer tissues
Subsequently, we explore the expression of miR-585-3p and the relationship between miR-585-3p and PSME3 in local tissues. As the Figure 5A shown, downregulated miR-585-3p in colon cancer was validated by qPCR analysis in our collected colon cancer tissue samples. Moreover, a negative correlation between miR-585-3p and PSME3 was obtained in our sets of colon cancer samples by Spearman correlation analysis (Figure 5B). Therefore, miR-585-3p is deregulated and shows a negative correlation to PSME3 in local colon cancer tissues.
Discussion
Recent studies have revealed the upregulation and oncogenic roles of PSME3 in several cancers as well as colon cancer.5,13 However, the underlying mechanism for PSME3 upregulation has not been explored. In the present study, we revealed for the first time that miR-585-3p can directly target PSME3 and suppress its pro-proliferation roles in colon cancer. Loss of miR-585-3p, at least partially, accounts for the molecular mechanism of PSME3 upregulation and associated oncogenic roles in colon cancer.
PSME3 upregulation has been reported in tumors of various tissue types including breast,8 pancreas,6 lung,11 thyroid7 and colon13 etc. Nevertheless, none of those studies have further presented the underlying mechanisms for its upregulation. We reveal that upregulated mRNA level of PSME3 is the primary reason for PSME3 overexpression in colon cancer tissues and cells indicating by online data and our experiment results. Generally, both transcriptional and post-transcriptional regulation mechanisms may regulate the level of mRNA. Transcriptional activation due to gene amplification, mutation, low methylated promoter, activation of upstream transcription factors, etc. is one of the common reasons for protein upregulation.28,29 Comparable promoter methylation status and hnRNA level eliminate the possibility of transcriptional mechanisms associated PSME3 upregulation. Post-transcriptional regulators, including miRNA, long no code RNA (lncRNA) and circle RNA (cirRNA), can also modulate mRNA level.16,29 Moreover, LncRNA and cirRNA usually serves as ceRNAs (competing endogenous RNAs) by sponging specific miRNAs and exerts its regulated roles.30 Therefore, miRNAs play vital roles in regulating protein level by inhibiting translation via directly targeting the mRNA for degradation and loss of tumor suppressed miRNAs are common causes of upregulated oncogenic proteins. Multiple miRNAs like miR-143, −145 can regulate colon cancer development via targeting KRAS and IRS-1, respectively.31,32 Apart from miR-7,23 which can target PSME3 in CHO cells, no other miRNAs targeting PSME3 have been confirmed yet. In our study, we reveal that miR-585-3p is a new negative regulator of PSME3 and loss of miR-585-3p, at least partially, accounts for the upregulation of PSME3 in colon cancer.
Recently, the tumor suppressive roles of miR-585 have been revealed by several groups. miR-585 downregulation, possibly caused by promoter hypermethylation, has been confirmed in gastric cancer,33 lung cancer,34 as well as oral squamous cell carcinoma.35 Accordingly, we also prove that miR-585-3p is also downregulated in colon cancer, suggesting miR-585 downregulation may be a universal event in carcinogenesis. Furthermore, restoration exhibits significant inhibitory on malignant characteristics involving proliferation, migration, and invasion. For example, miR-585 overexpression significantly suppresses the proliferation and migration of gastric cancer cells33 and non-small-cell lung cancer cells.34 Consistently, we indicate that miR-585-3p restoration also exhibits obviously inhibitory effects on proliferation and growth of colon cancer cells. Therefore, according to the existing research results proof, miR-585-3p mainly serves as a suppressor in tumorigenesis.
The functions of miRNAs are dependent on its downstream targets and the targets of a specific miRNA have tissue cell specificity.16,29 MAPK1 and hSMG-1 have been confirmed to be the targets of miR-585 and downregulated MAPK1 and hSMG-1, caused by miR-585 overexpression, leads to constrained cell proliferation and migration in gastric cancer and non-small-cell lung cancer, respectively.33,34 In this study, PSME3 is identified to be the target of miR-585-3p in colon cancer and ectopic PSME3 expression can almost antagonize the tumor suppressive effects of miR-585-3p on proliferation and growth of colon cancer cells. Moreover, a recent study has revealed that PSME3, serving as a new marker for the prognosis of colon cancer patients, is competent in promoting colon cancer progression by activating Hippo pathway and NF-κB pathway.13 Considering the inverse expression pattern between miR-585-3p and PSME3, miR-585-3p may also be useful in the prediction of prognosis of colon cancer patients.
Collectively, we reveal for the first time that downregulated miR-585-3p accounts for the PSME3 overexpression in colon cancer. What is more, we unfold that miR-585-3p restoration can significantly suppress cell proliferation and growth of colon cancer via negatively regulating PSME3 suggesting a new mechanism of carcinogenesis in colon cancer.
Acknowledgment
The present study was funded by the Natural Science Foundation of Hunan Province of China (no. 2017JJ2344).
Disclosure
There are no conflicts of interest for all authors in this work.
References
1. Pellino G, Warren O, Mills S, Rasheed S, Tekkis PP, Kontovounisios C. Comparison of Western and Asian guidelines concerning the management of colon cancer. Dis Colon Rectum. 2018;61(2):250–259. doi:10.1097/DCR.0000000000001012
2. Tamas K, Walenkamp AM, de Vries EG, et al. Rectal and colon cancer: not just a different anatomic site. Cancer Treat Rev. 2015;41(8):671–679. doi:10.1016/j.ctrv.2015.06.007
3. Xiao T, Zhu W, Huang W, et al. RACK1 promotes tumorigenicity of colon cancer by inducing cell autophagy. Cell Death Dis. 2018;9(12):1148. doi:10.1038/s41419-018-1113-9
4. Zhao Y, Hu X, Zuo X, Wang M. Chemopreventive effects of some popular phytochemicals on human colon cancer: a review. Food Funct. 2018;9(9):4548–4568. doi:10.1039/C8FO00850G
5. Mao I, Liu J, Li X, Luo H. REGγ, a proteasome activator and beyond? Cell Mol Life Sci. 2008;65(24):3971–3980. doi:10.1007/s00018-008-8291-z
6. Guo J, Hao J, Jiang H, et al. Proteasome activator subunit 3 promotes pancreatic cancer growth via c-Myc-glycolysis signaling axis. Cancer Lett. 2017;386:161–167. doi:10.1016/j.canlet.2016.08.018
7. Qi L, He W. REGgamma is associated with lymph node metastasis and T-stage in papillary thyroid carcinoma. Med Sci Monit. 2018;24:1373–1378. doi:10.12659/msm.905664
8. Yi Z, Yang D, Liao X, Guo F, Wang Y, Wang X. PSME3 induces epithelial-mesenchymal transition with inducing the expression of CSC markers and immunosuppression in breast cancer. Exp Cell Res. 2017;358(2):87–93. doi:10.1016/j.yexcr.2017.05.017
9. Chen H, Gao X, Sun Z, et al. REGgamma accelerates melanoma formation by regulating Wnt/beta-catenin signalling pathway. Exp Dermatol. 2017;26(11):1118–1124. doi:10.1111/exd.13394
10. Kontos CK. Surrogate prognostic biomarkers in OSCC: the paradigm of PA28gamma overexpression. EBioMedicine. 2015;2(8):784–785. doi:10.1016/j.ebiom.2015.07.032
11. He J, Cui L, Zeng Y, et al. REGgamma is associated with multiple oncogenic pathways in human cancers. BMC Cancer. 2012;12:75. doi:10.1186/1471-2407-12-75
12. Liu S, Zheng LL, Zhu YM, et al. Knockdown of REGgamma inhibits the proliferation and migration and promotes the apoptosis of multiple myeloma cells by downregulating NF-kappaB signal pathway. Hematology. 2018;23(5):277–283. doi:10.1080/10245332.2017.1385194
13. Wang Q, Gao X, Yu T, et al. REGgamma controls hippo signaling and reciprocal NF-kappaB-YAP regulation to promote colon cancer. Clin Cancer Res. 2018;24(8):2015–2025. doi:10.1158/1078-0432.CCR-17-2986
14. Qu JQ, Yi HM, Ye X, et al. MiR-23a sensitizes nasopharyngeal carcinoma to irradiation by targeting IL-8/Stat3 pathway. Oncotarget. 2015;6(29):28341–28356. doi:10.18632/oncotarget.5117
15. Monroig Pdel C, Chen L, Zhang S, Calin GA. Small molecule compounds targeting miRNAs for cancer therapy. Adv Drug Deliv Rev. 2015;81:104–116. doi:10.1016/j.addr.2014.09.002
16. Pekarsky Y, Croce CM. Noncoding RNA genes in cancer pathogenesis. Adv Biol Regul. 2019;71:219–223. doi:10.1016/j.jbior.2018.12.002
17. Wu K, Ma J, Zhan Y, et al. Down-regulation of MicroRNA-214 contributed to the enhanced mitochondrial transcription factor A and inhibited proliferation of colorectal cancer cells. Cell Physiol Biochem. 2018;49(2):545–554. doi:10.1159/000492992
18. Niu ZY, Li WL, Jiang DL, Li YS, Xie XJ. Mir-483 inhibits colon cancer cell proliferation and migration by targeting TRAF1. Kaohsiung J Med Sci. 2018;34(9):479–486. doi:10.1016/j.kjms.2018.04.005
19. Hu XX, Xu XN, He BS, et al. microRNA-485-5p functions as a tumor suppressor in colorectal cancer cells by targeting CD147. J Cancer. 2018;9(15):2603–2611. doi:10.7150/jca.24918
20. Sun Y, Xing X, Liu Q, et al. Hypoxia-induced autophagy reduces radiosensitivity by the HIF-1alpha/miR-210/Bcl-2 pathway in colon cancer cells. Int J Oncol. 2015;46(2):750–756. doi:10.3892/ijo.2014.2745
21. Knudsen KN, Lindebjerg J, Kalmar A, et al. miR-21 expression analysis in budding colon cancer cells by confocal slide scanning microscopy. Clin Exp Metastasis. 2018;35(8):819–830. doi:10.1007/s10585-018-9945-3
22. Wang J, Du Y, Liu X, Cho WC, Yang Y. MicroRNAs as regulator of signaling networks in metastatic colon cancer. Biomed Res Int. 2015;2015:823620.
23. Sanchez N, Gallagher M, Lao N, et al. MiR-7 triggers cell cycle arrest at the G1/S transition by targeting multiple genes including Skp2 and Psme3. PLoS One. 2013;8(6):e65671. doi:10.1371/journal.pone.0065671
24. Liu J, Huang W, Ren C, et al. Flotillin-2 promotes metastasis of nasopharyngeal carcinoma by activating NF-kappaB and PI3K/Akt3 signaling pathways. Sci Rep. 2015;5:11614. doi:10.1038/srep11614
25. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi:10.1016/s1476-5586(04)80047-2
26. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
27. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi:10.1093/nar/gkt1248
28. Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15(3):177–186. doi:10.1016/j.gpb.2016.12.005
29. Pu M, Chen J, Tao Z, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019 Feb;76(3):441–451. doi: 10.1007/s00018-018-2940-7
30. Wang Y, Hou J, He D, et al. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. 2016;32(4):211–224. doi:10.1016/j.tig.2016.02.001
31. Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28(10):1385–1392. doi:10.1038/onc.2008.474
32. Yu Y, Nangia-Makker P, Farhana L, Rajendra SG, Levi E, Majumdar AP. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer. 2015;14:98. doi:10.1186/s12943-014-0278-9
33. Hu L, Wu H, Wan X, et al. MicroRNA-585 suppresses tumor proliferation and migration in gastric cancer by directly targeting MAPK1. Biochem Biophys Res Commun. 2018;499(1):52–58. doi:10.1016/>j.bbrc.2018.03.116
34. Ding X, Yang Y, Sun Y, Xu W, Su B, Zhou X. MicroRNA-585 acts as a tumor suppressor in non-small-cell lung cancer by targeting hSMG-1. Clin Transl Oncol. 2017;19(5):546–552. doi:10.1007/s12094-016-1562-5
35. Uesugi A, Kozaki K, Tsuruta T, et al. The tumor suppressive microRNA miR-218 targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 2011;71(17):5765–5778. doi:10.1158/0008-5472.CAN-11-0368
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.