Back to Journals » Cancer Management and Research » Volume 12
Down-Regulation of Circ_0032833 Sensitizes Colorectal Cancer to 5-Fluorouracil and Oxaliplatin Partly Depending on the Regulation of miR-125-5p and MSI1
Received 30 June 2020
Accepted for publication 14 September 2020
Published 4 November 2020 Volume 2020:12 Pages 11257—11269
DOI https://doi.org/10.2147/CMAR.S270123
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Shouchao Li, Sheng Zheng
Department of Anorectal Surgery, Weifang People’s Hospital, Weifang 261000, People’s Republic of China
Correspondence: Sheng Zheng Email [email protected]
Background: 5-fluorouracil, leucovorin, and oxaliplatin (FOLFOX) is an effective chemotherapy for colorectal cancer (CRC) in clinic. It remains unclear regarding the effect of circular RNA (circRNA) circ_0032833 on regulating chemosensitivity in CRC.
Methods: Drug resistance analysis was performed by Cell Counting Kit-8 (CCK-8) assay. All RNA and protein levels were, respectively, measured via quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot. Cellular colony capacity, apoptosis and metastasis were evaluated using colony formation assay, Annexin-FITC/PI flow cytometry and transwell migration/invasion assays. The molecular combination was notarized using dual-luciferase reporter and RNA immunoprecipitation (RIP) assays. The in vivo experiment was conducted via xenograft tumors in mice.
Results: Circ_0032833 was significantly up-regulated in FOLFOX-resistant CRC and associated with drug resistance. Knockdown of circ_0032833 could sensitize FOLFOX-resistant CRC cells to 5-fluorouracil and oxaliplatin. Circ_0032833 was a miR-125-5p sponge, and miR-125-5p overexpression was responsible for the effect of circ_0032833 knockdown on 5-fluorouracil and oxaliplatin sensitivities. Besides, miR-125-5p targeted Musashi1 (MSI1) to increase the susceptibility of 5-fluorouracil and oxaliplatin in FOLFOX-resistant CRC cells. We found that circ_0032833 generated the regulation on MSI1 by sponging miR-125-5p. Circ_0032833 down-regulation also promoted the 5-fluorouracil and oxaliplatin sensitivities partly through the miR-125-5p/MSI1 axis in vivo.
Conclusion: This study illuminated an unambiguous mechanism circ_0032833/miR-125-5p/MSI1 on regulating 5-fluorouracil and oxaliplatin sensitivities in FOLFOX therapy, maybe providing a deep insight of resistance formation and developing a novel strategy to enhance chemosensitivity in CRC.
Keywords: circ_0032833, colorectal cancer, chemosensitivity, miR-125-5p, MSI1
Introduction
Colorectal cancer (CRC) is the third most common cancer (10.2% of the total cases), closely following the lung cancer (11.6) and breast cancer (11.6%) for men and women.1 5-fluorouracil, leucovorin, and oxaliplatin (FOLFOX) has been developed as the first-line chemotherapy, especially in metastatic or advanced and recurrent CRC.2–4 But the resistance of 5-fluorouracil and oxaliplatin usually results in a poor response to FOLFOX chemotherapy.5 To improve the anti-tumor response of FOLFOX therapy, knowing the occurrence mechanism of drug resistance and seeking effective biomarkers are imperative. As the well-known post-transcriptional regulators, non-coding RNAs (ncRNAs) have been recognized to mediate the chemoresistance of FOLFOX potentially.6
Non-coding circular RNAs (circRNAs) characterized by the specific covalent closed-loop without 5ʹ and 3ʹ free ends are stably enriched in many cancer cells as novel regulatory molecules.7 Recently, a number of differentially expressed circRNAs have been reported to be related to chemoresistant CRC.8 Hon et al have definitely identified that circ_0032833 was expressed with ectopic high level in FOLFOX-resistant CRC patient serum and cells.9 We will further explore the role of circ_0032833 on chemosensitivity in CRC.
MicroRNAs (miRNAs) mainly bind to 3ʹ untranslated regions (3ʹUTRs) of pre-messenger RNA to function as regulators in multiple cancers.10,11 Han et al found that miR-552 played a pivotal role in drug response of liver tumor,12 and abundant miRNAs have been used as diagnostic biomarkers for 5-fluorouracil chemoresistance in CRC.13 MiR-125 was a tumor inhibitor in CRC14 and its overexpression could promote the sensitivity of 5-fluorouracil in hepatocellular carcinoma cells.15 However, the effect of miR-125-5p on FOLFOX-resistant CRC is unclear.
Musashi1 (MSI1) is involved in various biological behaviors to regulate tumorigenesis, progression and chemoresistance.16 MSI1 was shown to enhance drug resistance in pediatric glioblastoma cells17 and non-small cell lung cancer cells.18 In addition, MSI1 could sensitize CRC cells to 5-fluorouracil.19 Here, the function of MSI1 on oxaliplatin sensitivity of CRC cells will also be addressed.
In different cancers, circRNAs can regulate gene expression via acting as “miRNAs sponges”.20 Thus, the relation among circ_0032833, miR-125-5p and MSI1 is set as another key point of our research.
Materials and Methods
Cell Culture and Establishment of FOLFOX-Resistant CRC Cells
HCT116 (a human CRC cell line) was bought from American Type Culture Collection (ATCC, Manassas, VA, USA), and maintained in McCoy’s 5A medium (Sigma-Aldrich, St. Louis, MO, USA) complemented with 10% fetal bovine serum (FBS; Sigma-Aldrich) and 1% antibiotic solution (10,000 unit/mL penicillin and 10 mg/mL streptomycin; Sigma-Aldrich). Temperature of 37°C, CO2 of 5% and 95% air were set as the standard conditions for HCT116 cell growth.
FOLFOX-resistant HCT116 (HCT116-R) cells were obtained by exposing the parental HCT-116 cells (HCT116-P) to 5-fluorouracil and oxaliplatin (5-FU and Oxa; Sigma-Aldrich) at clinically relevant doses as previously described,21 and finally cultivated in normal culture medium with a low dose of FOLFOX (5 µM 5-FU and 0.125 µM Oxa).
Drug Resistance Assay
Drug resistance was evaluated by the half inhibitory concentration (IC50) via cell viability assay in HCT116-P and HCT116-R cell lines treated with 5-FU or Oxa. Firstly, 3 × 103 cells or transfected cells were seeded into each well of 96-well plates to incubate for 24 h, followed by another 48 h incubation in presence of different concentrations of 5-FU (1 µM, 5 µM, 10 µM, 20 µM, 40 µM, 60 µM, 80 µM) and Oxa (1 µM, 2 µM, 5 µM, 10 µM, 20 µM, 40 µM, 60 µM). Whereafter, cells were added with 10 µL/well Cell Counting Kit-8 solution (Sigma-Aldrich) for 4 h, and the absorbance was determined by a microplate reader at 450 nm. The IC50 value was obtained according to the drug concentration at 50% cell viability.
Clinical Tissue Samples
Clinical specimens were approved to be used for this research by the Ethical Committee of Weifang People’s Hospital. Fifty CRC patients who have received FOLFOX therapy were finally subjected to surgical resection at Weifang People’s Hospital. According to the Response Evaluation Criteria In Solid Tumors (RECIST) criteria: complete response (CR), partial response (PR), stable disease (SD) and progressive disease (PD), these 50 patients were classified into Responders (n=25, CR+PR) and Nonresponders (n=25, SD+PD) toward FOLFOX therapy. All patients signed the written informed consent and the collected tissues were conserved in liquid nitrogen.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
Our RNA extraction was performed using Trizol (Beyotime, Shanghai, China). FastKing one-step reverse transcription kit (TIANGEN, Beijing, China) was used for complementary DNA (cDNA) synthesis and real-time PCR was implemented via FastFire qPCR PreMix (TIANGEN) in terms of the users’ manuals. As for data analysis, the comparative cycle threshold method (2−∆∆Ct) was used for calculating the relative expression levels. The used housekeeping genes were glyceraldehyde-3-phosphate dehydrogenase (GAPDH; for circ_0032833 or MSI1) and U6 (for miR-125-5p). All forward (F) and reverse (R) primer sequences were presented as below: circ_0032833: 5ʹ-CATCAATGCTGAGGAGCAGA-3ʹ (F) and 5ʹ-GCCAAGGGCAGTACTCAGAA-3ʹ (R); miR-125-5p: 5ʹ-GGGTCCGAGGTATTCGCACT-3ʹ (F) and 5ʹ-TCCCTGAGACCCTTTAACCTGTG-3ʹ (R); MSI1: 5ʹ-GCGACACTGCTGGACAGGAATTA-3ʹ (F) and 5ʹ-AGAGGGACACACAGAAGGGGGAT-3ʹ (R); GAPDH: 5ʹ-AGAAGGCTGGGGCTCATTTG-3ʹ (F) and 5ʹ-AGGGGCCATCCACAGTCTTC-3ʹ (R); U6: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (F) and 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ (R).
Western Blot
Cells in 24-well plates were washed with ice-cold phosphate buffer solution (PBS; Beyotime) and the protein was extracted from the harvested cell pellets using radio-immunoprecipitation assay buffer (RIPA; Beyotime) added with Phenylmethanesulfonyl fluoride (PNSF; Beyotime). Human tissues were cut into small pieces and sufficiently lysed in RIPA+PMSF solution, followed by the centrifugation at 12,000 g for 5 min to obtain the supernatant protein. Next, the separation of protein from cells or tissues was conducted by the electrophoresis on TruPAGE™ Precast Gels (12%, 10 × 10 cm, 12-well; Sigma-Aldrich) for 120 min and separated proteins were electrically moved onto the PVDF Transfer Membrane (0.2 µm; Beyotime). After immersing in 5% (w/v) non-fat milk (Beyotime) in tris-buffered saline with 0.1% Tween-20 (TBST) overnight, the membranes were incubated in primary antibodies diluted with TBST at room temperature for 4 h and then washed in TBST for three times (5 min/time). The membranes were incubated with diluted secondary antibody for 1 h and washed in TBST, then ECL Substrate Kit (Abcam, Cambridge, UK) and ImageLab software version 4.1 (Bio-Rad, Hercules, CA, USA) were applied for analyzing protein density. The antibodies (Abcam) and dilutability were as follows: anti-multidrug resistance-associated protein 1 (anti-MRP-1; ab234098, 1:1000), anti-myeloid cell leukemia-1 (anti-MCL-1; ab32087, 1:1000), anti-caspase-3 (ab44976, 1:1000), anti-N-cadherin (anti-N-cad; ab18203, 1:1000), anti-E-cadherin (anti-E-cad; ab15148, 1:1000), anti-MSI1 (ab21628, 1:1000), anti-GAPDH (ab9485, 1:2500) and Goat Anti-Rabbit IgG H&L (HRP) (ab205718, 1:5000).
Cell Transfection
The small interfering RNA (siRNA), short hairpin RNA (shRNA) and miRNA-related oligonucleotides were all purchased from GenePharma (Shanghai, China): siRNA targeting circ_0032833 (si-circ), shRNA targeting circ_0032833 (sh-circ), miR-125-5p mimic, miR-125-5p inhibitor and corresponding negative controls (si-NC, sh-NC, mimic NC and inhibitor NC). The overexpression vector of MSI1 (oe-MSI1) was bought from GENEWIZ (Suzhou, China). Cell transfection was then administrated using Lipofectamine3000 reagent (Invitrogen, Carlsbad, CA, USA) as per the provided instruction book.
Colony Formation Assay
3 × 102 cells in 2 mL complete media were seeded into the 6-well plates, and cultured in standard growth condition for two weeks. After macroscopic colonies were fixed and stained in 4% paraformaldehyde (Beyotime) and crystal violet (Beyotime), 2 mL PBS was exploited for washing out excess dye and the clonal number was counted.
Annexin V-FITC/PI Flow Cytometry
Annexin-FITC Apoptosis detection kit (Solarbio, Beijing, China) was used to examine apoptotic cells, referring to the producer’s specification. Briefly, 100 µL cell suspension in 1 × Binding Buffer (1 × 105 cells) was pipetted into a 2-mL tube, followed by adding 5 µL Annexin V-FITC for 10 min and propidium iodide (PI) for 5 min in the dark. Afterwards, the tube was supplemented with PBS to 500 µL and the apoptotic cells were instantly detected via the flow cytometer (BD Biosciences, San Diego, CA, USA). The (Annexin V+/PI-)-labelled cells were considered as early apoptotic cells and (Annexin V+/PI+)-labelled cells were as late apoptotic cells. Then, cell apoptosis rate (apoptotic cells/total cells × 100%) was calculated.
Transwell Assay
A transwell chamber (Corning Inc., Corning, NY, USA) was used for migration determination while the matrigel (Corning Inc.)-coated chamber was applied to analyze invasion ability. Harvested cells were resuspended in medium without serum, and added to the upper chamber. The migrated or invaded phenomenon could occur after adding the medium containing 10% FBS into the lower chamber. After cells passed into the lower chamber were fastened by 4% paraformaldehyde and dyed in crystal violet, the images of migrated and invaded cells were obtained by an inverted microscope (Olympus, Tokyo, Japan) and the number was counted.
Dual-Luciferase Reporter Assay
The wild-type (wt) sequences of circ_0032833 and MSI1 3ʹUTR were respectively cloned into the pGL3 luciferase empty vector (Promega, Madison, WI, USA) to construct circ_0032833 wt and MSI1 3ʹUTR wt luciferase plasmids. After the miR-125-5p binding sites in wt sequences of circ_0032833 and MSI1 3ʹUTR were mutated, the mutant-type (mut) sequences were used to obtain the novel plasmids circ_0032833 mut and MSI1 3ʹUTR mut. These constructed plasmids were respectively transfected into 293T cells (COBIOER, Nanjing, China) in Dulbecco’s modified eagle medium (DMEM; Sigma-Aldrich), accompanied with miR-125-5p mimic or mimic NC. 48 h later, cells were collected and the relative luciferase activity (ratio of firefly and renilla) was examined via the dual luciferase reporter system (Promega).
RNA Immunoprecipitation (RIP) Assay
Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was employed to analyze the interplay between circ_0032833 and miR-125-5p. After cell lysis in RIP lysis buffer for 1 h, cell suspension was incubated with A/G (anti-Ago2 or anti-IgG)-covered magnetic beads overnight at 4°C. The precipitated RNA-binding protein complexes were washed from magnetic beads, and qRT-PCR was used for circ_0032833 and miR-125-5p analysis following the protein removal and RNA acquisition.
In vivo Experiment
We purchased 24 BALB/c nude mice (6-week-old) from Shanghai SIPPR-BK Laboratory Animal Co. Ltd. (Shanghai, China) and randomly divided them into four groups: sh-NC+5-FU, sh-circ+5-FU, sh-NC+Oxa and sh-circ+Oxa. Mice were subcutaneously injected with 5×106 HCT116-R cells transfected sh-circ or sh-NC lentiviral vector. After 7 d, tumor volume (length × width2 × 0.5) was measured by digital caliper and mice were subjected to intraperitoneal injection with 50 mg/kg 5-FU or 10 mg/kg Oxa twice a week. Then, tumor volume was recorded per 5 d, and all mice were euthanatized using CO2 asphyxia method at 32 d. The excised tumors were weighed and used for RNA or protein extraction, followed by the detection of related RNA and proteins. This experiment was administrated based on the ratification from the Animal Ethical Committee of Weifang People’s Hospital. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
Our assays were all independently performed three times. Data expressed as mean ± standard deviation (SD) were carefully processed applying with SPSS 22.0 and Graphpad prism 7. The intergenic linear relationship was analyzed using Spearman correlation coefficient. Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s test were applied for difference comparison. Difference was statistically significant when P value <0.05.
Results
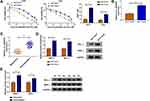
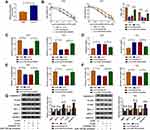
Through cell viability analysis, we found that IC50 values of 5-FU (40 µM) and Oxa (22 µM) in HCT116-R cells were much higher than HCT116-P cells, confirming that HCT116-R cells were resistant to 5-FU and Oxa (Figure 1A). The qRT-PCR indicated that circ_0032833 expression was prominently increased in HCT116-R cells and clinical Nonresponders compared with HCT116-P cells and Responders (Figure 1B-C). Moreover, Western blot demonstrated that multidrug resistance-related proteins MRP-1 and MCL-1 were signally up-regulated in HCT116-R cells (Figure 1D) and Nonresponders (Figure 1E) in contrast with their respective control group. These data collectively suggested that circ_0032833 was closely related to drug resistance in FOLFOX-resistant CRC tissues and cells.
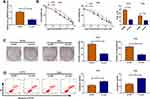
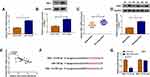
In HCT116-R cells, we used si-circ transfection to knock down the expression of circ_0032833 (Figure 2A). In si-circ-transfected HCT116-R cells, CCK-8 revealed that the viable cell number was overtly declined with the increase of 5-FU and Oxa, then the IC50 values of 5-FU and Oxa were found to be lessened compared to si-NC-transfected HCT116-R cells (Figure 2B). Colony formation assay and flow cytometry showed that circ_0032833 knockdown triggered the decreased colonies (Figure 2C) and increased apoptosis rate (Figure 2D) in HCT116-R cells treated with 30 µM 5-FU or Oxa. Determination of cell migration and invasion (Figure 3A-B) exhibited the obvious inhibition in si-circ group contrasted with si-NC group. By analyzing the related protein levels, we also discovered the si-circ transfection downregulated the protein expression of multidrug resistance-promoting markers (MRP-1 and MCL-1) and pro-metastasis marker (N-cad) but stimulated the levels of anti-metastasis marker (E-cad) and apoptosis positive marker (caspase-3) in 5-FU or Oxa-treated HCT116-R cells (Figure 3C). All these results proved that circ_0032833 down-regulation increased 5-fluorouracil and oxaliplatin sensitivities to inhibit the progression of FOLFOX-resistant CRC cells.
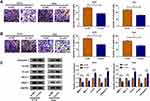
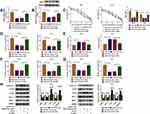
Starbase3.0 has predicted that 20 miRNAs had the binding sites for circ_0032833 and 3 miRNAs (miR-506-3p, miR-124-3p, miR-125-5p) that were downregulated in CRC were selected as the candidate targets for circ_0032833. Our screening assays indicated that three miRNAs were all downregulated in Nonresponders (n=5) compared to Responders (n=5) and their levels were significantly increased by knockdown of circ_0032833 (Supplemental Figure 1A-B). Thus, miR-125-5p with most obvious upregulation was chosen for further research. Then, we found the downregulation of miR-125-5p not only in HCT116-R cells but also in 25 Nonresponders, in comparison with HCT116-P cells and 25 Responders (Figure 4A-B). Interestingly, circ_0032833 was negatively correlated to miR-125-5p (r=−0.5273, P=0.0068) in Nonresponsive tissues (Figure 4C). Starbase 3.0 presented the theoretical binding between the sequences of circ_0032833 and miR-125-5p (Figure 4D), and then we performed the dual-luciferase reporter assay in 293T cells to validate that miR-125-5p could interact with circ_0032833 wt not circ_0032833 mut (Figure 4E). As RIP analysis in Figure 4F, both circ_0032833 and miR-125-5p were largely captured by anti-Ago2 compared to anti-IgG, indicating the direct interaction between circ_0032833 and miR-125-5p.
Compared to si-NC transfection, si-circ transfection apparently heightened the relative level of miR-125-5p (Figure 5A). Given that circ_00328333 could act as a miR-125-5p sponge, we further researched their relation on regulating chemosensitivity in HCT116-R cells. As Figure 5B depicted, miR-125-5p inhibitor significantly abated the inhibitory effects of si-circ on IC50 of 5-FU and Oxa. By comparison with si-circ+inhibitor NC group, the colony formation capacity in si-circ+miR-125-5p inhibitor group was enhanced (Figure 5C) while cell apoptosis was restrained on the contrary (Figure 5D). Similarly, migrated and invaded cells were increased after co-transfection of si-circ and miR-125-5p inhibitor relative to alone si-circ transfection (Figure 5E-F). Western blot also testified that the si-circ-induced inhibition on resistance, metastasis and promotion on apoptosis were all relieved following the down-regulation of miR-125-5p in HCT116-R cells treated of 5-FU and Oxa (Figure 5G). Taken together, miR-125-5p inhibitor reversed the stimulative effects of circ_0032833 knockdown on 5-fluorouracil and oxaliplatin sensitivities.
MSI1 Was a Target of miR-125-5p
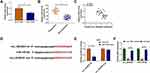
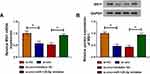
As for the target genes of miR-125-5p, many potential targets (>3000) were predicted by online Starbase3.0 and some genes (XIAP, FOXQ1, USP22, ABCC1, NOTCH-1, MSI1) have been reported to be implicated in the chemoresistance of CRC. The qRT-PCR results have indicated their upregulation in Nonresponders contrasted with Responders and Western blot showed that miR-125-5p mimic resulted in most evident downregulation of the protein expression of MSI1 (Supplemental Figure 2A-B). Then we further researched the target relation between miR-125-5p and MSI1 in CRC. Through the analysis of qRT-PCR and Western blot, MSI1 was identified as an up-regulated gene in HCT116-R cells (Figure 6A-B), as well as in Nonresponders (Figure 6C-D). Spearman correlation coefficient displayed a negative linear relationship (r=−0.5689, p=0.003) between miR-125-5p and MSI1 mRNA levels (Figure 6E). In Starbase 3.0 online software, we found the hypothetic binding sites of miR-125-5p in the MSI1 3ʹUTR sequence (Figure 6F); subsequently, the actual combination between miR-125-5p and MSI1 3ʹUTR was confirmed by the decrease of the relative luciferase activity in miR-125-5p mimic+MSI1 3ʹUTR wt co-transfection group (Figure 6G). Therefore, MSI1 can be used as a downstream target of miR-125-5p.
MiR-125-5p Worked as a Sensitizer of 5-Fluorouracil and Oxaliplatin in FOLFOX-Resistant CRC Cells via Targeting MSI1
By designing the four transfection groups: mimic NC, miR-125-5p mimic, miR-125-5p mimic+vector and miR-125-5p mimic+oe-MSI1, the role of miR-125-5p on drug sensitivity and how it acted would be addressed. As shown in Figure 7A-B, miR-125-5p mimic repressed the mRNA and protein levels of MSI1 in HCT116-R cells while oe-MSI1 transfection promoted MSI1 expression of miR-125-5p mimic group in a revertible manner. CCK-8 assay indicated that the decreased IC50 values of 5-FU and Oxa in miR-125-5p mimic group were elevated in miR-125-5p mimic+oe-MSI1 group, hinting that miR-125-5p inhibited 5-FU and Oxa resistance by targeting MSI1 (Figure 7C). In HCT116-R cells treated with 30 µM 5-FU or Oxa, we found that miR-125-5p overexpression led to the suppression of colony formation ability (Figure 7D) and the increased apoptosis rate (Figure 7E) along with the blockage of migration/invasion (Figure 7F-G), which were all restored after the up-regulation of MSI1. Meanwhile, miR-125-5p mimic down-regulated the levels of MRP-1, MCL-1 and N-cad proteins but motivated the expression of E-cad and caspase-3, whereas the introduction of oe-MSI1 vector abrogated the miR-125-5p-induced resistance and metastasis obstruction as well as apoptosis promotion (Figure 7H). Above works manifested the role of miR-125-5p as a sensitizer of 5-fluorouracil and oxaliplatin in FOLFOX-resistant CRC cells by targeting MSI1.
Owing to the miR-125-5p link between circ_0032833 and MSI1, the reverted transfection was performed to analyze whether circ_0032833 could affect MSI1 via miR-125-5p. As Figure 8A-B illustrated, si-circ transfection evidently repressed the MSI1 mRNA and protein expression in HCT116-R cells, but the inhibitory effects were offset following the down-regulation of miR-125-5p. Indirectly, circ_0032833 could regulate MSI1 through directly binding to miR-125-5p.
Xenograft models were used for our experiment in vivo. After injection of HCT116-R cells and treatment of 5-FU, we found that tumor volume was reduced with the increase of time (Figure 9A) and the finally excised tumors were lighter (Figure 9B) in sh-circ group by comparing with sh-NC group. The expression levels of circ_0032833 and MSI1 (mRNA and protein) were lower while miR-125-5p was higher in sh-circ group than these in sh-NC group (Figure 9C-D). Likewise, the interference of circ_0032833 expression also decreased tumor volume and weight (Figure 9E-F) by the miR-125-5p/MSI1 axis (Figure 9G-H) in Oxa-treated mice. Additionally, sh-circ introduction evoked the resistance inhibition (the down-regulated MRP-1 and MCL-1), metastasis impairment (the up-regulated E-cad and down-regulated N-cad) and apoptosis enhancement (the increased caspase-3) in tumors from 5-FU or Oxa-treated mice (Figure 9I). Also in vivo, circ_0032833 knockdown contributed to the 5-fluorouracil and oxaliplatin sensitivities via the miR-125-5p/MSI1 axis.
Discussion
Chemoresistance has become an intractable issue in clinical CRC treatment, which makes it increasingly important to facilitate chemosensitivity. In the current report, knockdown of circ_0032833 was proved to contribute to the sensitivity of CRC cells to 5-fluorouracil and oxaliplatin via regulating MSI1 expression by targeting miR-125-5p.
CircRNAs are now in the spotlight as potential regulators in human cancers, also in resistance of cancer chemotherapy.22 Liu et al discovered that circHIPK3 expedited chemoresistance of gemcitabine in pancreatic cancer cells23 and Yang et al clarified that the low circ-CDR1as had promotive effect on chemosensitivity in 5-fluorouracil-resistant breast cancer cells.24 Microarray analysis of circRNAs affirmed their dysregulation in 5-fluorouracil-based chemoradiation-resistant CRC cells.25 And the latest study has exhibited that circRNA ciRS-122 induced the generation of resistance in oxaliplatin-sensitive CRC cells.26 Circ_0032833 was up-regulated in FOLFOX-resistant CRC cells by our qRT-PCR detection. Functionally, siRNA-mediated circ_0032833 interference notably decreased resistance both of 5-fluorouracil and oxaliplatin along with cancer retardation by analyzing the cellular process. Thus, the repression of circ_0032833 could sensitize FOLFOX-resistant CRC cells to 5-fluorouracil and oxaliplatin in accordance with the above studies.
Subsequently, miR-125-5p was found to be down-regulated in FOLFOX-resistant CRC (tissues and cells) and negatively related to circ_0032833 level. Growing researches have demonstrated the miRNA sponge mechanism of circRNAs in all types of tumor cells. For instance, circ_0001564 acted as a oncogene to increase proliferation and inhibit apoptosis of osteosarcoma cells through sequestering miR-29c-3p;27 circular RNA profiling proved circADAMTS13 as a miR-484 sponge to be an inhibitor in the progression of hepatocellular carcinoma cells;28 circ_0000523 also was reported to regulate CRC development by relying on the sponge of miR-31.29 It was similar that miR-125-5p also could be sponged by circ_0032833, and the chemosensitivity regulation of circ_0032833 knockdown on FOLFOX-resistant CRC cells was achieved by the elevation of miR-125-5p.
MSI1 was an overexpressed gene in CRC tissues and cells with FOLFOX-resistance through our expression analysis of qRT-PCR and Western blot. Like circ_0032833 and miR-125-5p, miR-125-5p also could directly target MSI1. miR-125-5p was identified as a great promoter in sensitizing CRC cells to 5-fluorouracil and oxaliplatin, by combining with the 3ʹUTR of MSI1 to block MSI1 expression. Moreover, MSI1 level was regulated by the circ_0032833-mediated miR-125-5p change here. Zhang et al have declared that circ_0063809 boosted the paclitaxel resistance in ovarian cancer via sponging miR-1252 to up-regulate FOXR2.30 Sang et al showed that circ_0025202 enhanced tamoxifen efficacy of breast cancer cells through affecting the miR-182-5p/FOXO3a axis.31 Herein, circ_0032833 down-regulation regulated the miR-125-5p/MSI1 axis to elevate 5-fluorouracil and oxaliplatin efficacies of CRC not only in vitro but also in vivo.
Conclusion
In summary, the present study identified circ_0032833/miR-125-5p/MSI1 axis as a novel 5-fluorouracil and oxaliplatin resistance mechanism in CRC, and circ_0032833 as a diagnostic target for resistance generation. Promisingly, the efficiency of FOLFOX chemotherapy will be improved by increasing the 5-fluorouracil and oxaliplatin susceptibilities after knocking down circ_0032833.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Venook AP, Niedzwiecki D, Lenz HJ, et al. Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: a randomized clinical trial. JAMA. 2017;317(23):2392–2401. doi:10.1001/jama.2017.7105
3. Liang XB, Hou SH, Li YP, Wang LC, Zhang X, Yang J. Irinotecan or oxaliplatin combined with 5-fluorouracil and leucovorin as first-line therapy for advanced colorectal cancer: a meta-analysis. Chin Med J (Engl). 2010;123(22):3314–3318.
4. Baba H, Hayashi N, Emi Y, et al. A multicenter Phase II clinical study of oxaliplatin, folinic acid, and 5-fluorouracil combination chemotherapy as first-line treatment for advanced colorectal cancer: a Japanese experience. Surg Today. 2011;41(12):1610–1616. doi:10.1007/s00595-011-4589-9
5. Li P, Zhang X, Wang H, et al. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol Cancer Ther. 2017;16(4):739–751. doi:10.1158/1535-7163.MCT-16-0591
6. Hon KW, Abu N, Ab Mutalib NS, Jamal R. miRNAs and lncRNAs as predictive biomarkers of response to FOLFOX therapy in colorectal cancer. Front Pharmacol. 2018;9:846. doi:10.3389/fphar.2018.00846
7. Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015–5021. doi:10.7150/jca.30828
8. Abu N, Hon KW, Jeyaraman S, et al. Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics. 2019;11(8):875–884. doi:10.2217/epi-2019-0042
9. Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal R, Abu N. Extracellular vesicle-derived circular RNAs confers chemoresistance in colorectal cancer. Sci Rep. 2019;9(1):16497. doi:10.1038/s41598-019-53063-y
10. Zheng J, Zhang Y, Cai S, et al. MicroRNA-4651 targets bromodomain-containing protein 4 to inhibit non-small cell lung cancer cell progression. Cancer Lett. 2020;476:129–139. doi:10.1016/j.canlet.2020.02.018
11. Weng Q, Chen M, Yang W, et al. Integrated analyses identify miR-34c-3p/MAGI3 axis for the Warburg metabolism in hepatocellular carcinoma. FASEB J. 2020. doi:10.1096/fj.201902895R
12. Han T, Zhang Y, Yang X, et al. miR-552 regulates liver tumor-initiating cell expansion and sorafenib resistance. Mol Ther Nucleic Acids. 2020;19:1073–1085. doi:10.1016/j.omtn.2019.12.043
13. Zhang X, Zhang H, Shen B, Sun XF. Novel MicroRNA biomarkers for colorectal cancer early diagnosis and 5-fluorouracil chemotherapy resistance but not prognosis: a study from databases to AI-assisted verifications. Cancers (Basel). 2020;12(2). doi:10.3390/cancers12020341
14. Yang M, Tang X, Wang Z, Wu X, Tang D, Wang D. miR-125 inhibits colorectal cancer proliferation and invasion by targeting TAZ. Biosci Rep. 2019;39(12). doi:10.1042/BSR20190193
15. Jiang JX, Gao S, Pan YZ, Yu C, Sun CY. Overexpression of microRNA-125b sensitizes human hepatocellular carcinoma cells to 5-fluorouracil through inhibition of glycolysis by targeting hexokinase II. Mol Med Rep. 2014;10(2):995–1002. doi:10.3892/mmr.2014.2271
16. Kudinov AE, Karanicolas J, Golemis EA, Boumber Y. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin Cancer Res. 2017;23(9):2143–2153. doi:10.1158/1078-0432.CCR-16-2728
17. Potschke R, Gielen G, Pietsch T, et al. Musashi1 enhances chemotherapy resistance of pediatric glioblastoma cells in vitro. Pediatr Res. 2019. doi:10.1038/s41390-019-0628-9
18. Lang Y, Kong X, He C, et al. Musashi1 promotes non-small cell lung carcinoma malignancy and chemoresistance via activating the Akt signaling pathway. Cell Physiol Biochem. 2017;44(2):455–466. doi:10.1159/000485012
19. Chiou GY, Yang TW, Huang CC, et al. Musashi-1 promotes a cancer stem cell lineage and chemoresistance in colorectal cancer cells. Sci Rep. 2017;7(1):2172. doi:10.1038/s41598-017-02057-9
20. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. doi:10.1007/978-981-13-1426-1_6
21. Yu Y, Kanwar SS, Patel BB, Nautiyal J, Sarkar FH, Majumdar AP. Elimination of colon cancer stem-like cells by the combination of curcumin and FOLFOX. Transl Oncol. 2009;2(4):321–328. doi:10.1593/tlo.09193
22. Jeyaraman S, Hanif EAM, Ab Mutalib NS, Jamal R, Abu N. Circular RNAs: potential regulators of treatment resistance in human cancers. Front Genet. 2019;10:1369. doi:10.3389/fgene.2019.01369
23. Liu Y, Xia L, Dong L, et al. CircHIPK3 promotes Gemcitabine (GEM) resistance in pancreatic cancer cells by sponging miR-330-5p and targets RASSF1. Cancer Manag Res. 2020;12:921–929. doi:10.2147/CMAR.S239326
24. Yang W, Gu J, Wang X, et al. Inhibition of circular RNA CDR1as increases chemosensitivity of 5-FU-resistant BC cells through up-regulating miR-7. J Cell Mol Med. 2019;23(5):3166–3177. doi:10.1111/jcmm.14171
25. Xiong W, Ai YQ, Li YF, et al. Microarray analysis of circular RNA expression profile associated with 5-fluorouracil-based chemoradiation resistance in colorectal cancer cells. Biomed Res Int. 2017;2017:8421614. doi:10.1155/2017/8421614
26. Wang X, Zhang H, Yang H, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–555. doi:10.1002/1878-0261.12629
27. Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495(3):2369–2375. doi:10.1016/j.bbrc.2017.12.050
28. Qiu L, Huang Y, Li Z, et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13(2):441–455. doi:10.1002/1878-0261.12424
29. Jin Y, Yu LL, Zhang B, Liu CF, Chen Y. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res. 2018;51(12):e7811. doi:10.1590/1414-431X20187811
30. Zhang S, Cheng J, Quan C, et al. circCELSR1 (hsa_circ_0063809) contributes to paclitaxel resistance of ovarian cancer cells by regulating FOXR2 expression via miR-1252. Mol Ther Nucleic Acids. 2019;19:718–730. doi:10.1016/j.omtn.2019.12.005
31. Sang Y, Chen B, Song X, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27(9):1638–1652. doi:10.1016/j.ymthe.2019.05.011
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.