Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 13
Does the Autophagy Related Gene 7 (ATG7) Have a Role in Non-Melanoma Skin Cancer?
Authors Samaka RM, Basha MA, Mansour E
Received 5 July 2019
Accepted for publication 29 November 2019
Published 20 January 2020 Volume 2020:13 Pages 49—58
DOI https://doi.org/10.2147/CCID.S222051
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Rehab M Samaka, 1 Mohammed A Basha, 2 Eman Mansour 3
1Pathology Department, Faculty of Medicine, Menoufia University, Al Minufya, Egypt; 2Dermatology, Andrology and STDs Department, Faculty of Medicine, Menoufia University, Al Minufya, Egypt; 3Ministry of Health, El Menshawy General Hospital, Tanta, Egypt
Correspondence: Eman Mansour
Ministry of Health, El Menshawy General Hospital, Tanta, Egypt
Tel +201273906558
Fax +20403305158
Email [email protected]
Rehab M Samaka
Pathology Department, Faculty of Medicine, Menoufia University, Al Minufya, Egypt
Tel +201002806239
Fax +20482317508
Email [email protected]
Purpose: To evaluate the role of autophagy related gene 7 (ATG7) in non-melanoma skin cancer.
Subjects and Methods: This retrospective and prospective case-control study was performed on 104 patients with non-melanoma skin cancer (NMSC) in addition to 20 apparently healthy subjects matched for age and sex as a control group. Multiple skin biopsies were taken for immunohistochemical evaluation of ATG7 expression.
Results: Both epithelial and stromal ATG7 were expressed in all participants while all patients showed nucleocytoplasmic localization and controls showed both cytoplasmic and nucleocytoplasmic expression. In addition, significantly higher H-scores of ATG7 in both epithelium and stroma were detected in patients compared to controls (P< 0.001).
Conclusion: ATG7 nucleocytoplasmic topographic localization might be involved in the pathogenesis of NMSC, which can open the gate for new target therapy for this skin cancer.
Keywords: autophagy, autophagy related gene 7, BCC, SCC, NMSC
Introduction
Although malignant melanoma (MM) is more lethal than other kinds of skin cancer, non-melanoma skin cancer (NMSC) – including basal cell carcinoma (BCC) and squamous cell carcinoma (SCC) – has a greater lethality and is also associated with substantial morbidity, loss of function, disfigurement, and costs.1,2 The incidence of NMSC has been increasing over the past decades since between 2 and 3 million NMSC occur globally each year.3,4
The Pathogenesis of BCC and SCC is usually combined with environment factors, mainly ultraviolet (UV) irradiation by long-term sun exposure. Different environmental factors are usually involved in the pathogenesis of BCC and SCC, mainly ultraviolet (UV) irradiation through long-term sun exposure.
Ultraviolet light can induce DNA damage randomly in keratinocytes. Also it can mutate genes that are crucial for skin epidermis control and monitoring. NMSC most often grows on regions of skin that are frequently subjected to the sun.5
Autophagy is a cellular process that retains the homeostasis and integrity of cells and tissues through the degradation of senescent, defective subcellular organs, infectious agents, and misfolded proteins.6 In recent years, studies have shown that autophagy is a vital arbiter of cell fate decisions and plays an important role in inflammation, pathogen clearance, and antigen presentation. This pathway is also essential for maintaining cellular homeostasis in the skin.7
Autophagy is suggested to have both tumor-suppressing and tumor-promoting functions during cancer progression. This functional duality was recently reported to be due to its association with diverse oncogenes or tumor suppressors.8 Generally, autophagy has a tumor suppressor role in normal cells but acts as a survival mechanism for established tumors.9 Squamous cell carcinoma and melanoma have demonstrated elevated concentrations of autophagic activity in which tumor aggressiveness is associated with elevated autophagic activity.10
Recent studies indicate that chloroquine inhibition of autophagy could enhance cell death in metastatic SCC cells caused by flavonoid luteolin.11
Autophagy related protein 7 (ATG7) is a main regulator for autophagosome formation and one of the most significant ATG members on the basis of which many animal models are constructed and developed to explore the role of autophagy in controlling physiological procedures in cells.12 Different studies were designed to assess the role of autophagy related protein 7 in human cancers. Autophagy inhibition in intestinal epithelial cells by conditional inactivation of ATG7 inhibits the development of precancerous lesions in patients who are at great risk of developing colorectal cancer, ATG7 deficiency leading to p53-mediated cell-cycle arrest in tumor cells but not in normal tissue.13 Also it was reported that ATG7 deficiency can increase breast cancer cells resistance to photodynamic therapy.14
This study was designed to evaluate the role of autophagy related protein 7 (ATG7) in NMSC.
Patients and Methods
This retrospective and prospective case-control study was carried out on 104 patients with NMSC (77 patients with BCC, 27 patients with SCC) and 20 apparently healthy volunteers’ age and gender matched as a control. Prospective cases were selected from Outpatient Clinics of Dermatology and Plastic Surgery at Menoufia University Hospital. All participants provided written informed consent before enrollment in the study.
For the retrospective part of the study, tissue blocks were collected from archives of the Pathology Department, Faculty of Medicine, Menoufia University, Egypt based on the institutional review board’s decision regarding patient consent spanning the period between April 2018 and January 2019. The study was approved by the Ethical Committee of Menoufia University and was conducted in accordance with the Declaration of Helsinki.
Prospective cases were subjected to a full history taking, general, and dermatological examinations, while retrospective cases were collected from patients’ files and their data were maintained with confidentiality.
Skin Biopsies
For prospective patients, full-thickness excision of the lesion with a minimum of 5-mm clear margins was carried out for all patients.15 The skin biopsies were preserved in 10% neutral buffered formalin then subjected to the routine histopathological examination. From each block, several 5 micron (5 µm) thick sections were taken and stained with hematoxylin and eosin (H&E) stain for the routine histopathologic evaluation.16 Sections on positive charged slides were immune histochemical (IHC) stained of ATG7. Assessment of ATG7 IHC stained slides in both epithelium and stroma (as staining state: either positive or negative, staining localization.17 Histo-score (H score) was used to evaluate the positive cases, where both the intensity and the percentage of positivity were measured using the following formula: H score=+3 (strong intensity)×%+2 (moderate intensity)×%+1 (mild intensity)×%. The resulting score ranges between 0 and 300.18
Immunohistochemical Staining
Immunohistochemical staining was done using rabbit anti-ATG7 polyclonal antibody (Cat. # GTX32459, Gene Tex, CA; isotype IgG; diluted at 1:100). All slides were de-paraffinized with xylene and subsequently rehydrated at reduced ethanol concentration. Antigen retrieval by microwave heating (20 minutess; 10 mmol/citrate buffer, pH 6.0) after inhibition of endogenous peroxidase activity (15 minutess of hydrogen peroxidase) was performed. The primary antibodies were applied on the slides and incubated all the night at room temperature in a humidity chamber. Finally, the detection of bound antibody was achieved using a modified labeled avidin–biotin (LAB) reagent for 20 minutes then a phosphate buffered saline (PBS) wash. A concentration of 0.1% solution of diaminobenzidine (DAB) was used for 5 minutes as a chromogen. Slides were further counterstained with Mayer’s hematoxylin for 5–10 minutes. The preparation of negative control slides was done by omitting the primary antibodies from the staining procedure. Normal human gastric tissues were used as a positive control for ATG7.
Statistical Analysis
Data were collected, tabulated, and statistically analyzed using a personal computer with “Statistical Package for the Social Sciences (SPSS) version 22“ program. Qualitative data is expressed as both number and percentage. Quantitative data is expressed as: arithmetic mean (X), standard deviation (SD), percentage (%), and median. Chi-square test (χ2-test) was used for comparing qualitative variables. Student’s t-test and Mann–Whitney U-test (U-test) were used for comparing quantitative variables. Kruskal Wallis test was used for testing the equality of population medians among groups, as it is an extension of the Mann–Whitney test. One-way Anova test (F) was used for comparison between three or more quantitative variables. Pearson correlation was used to study the correlation between parametric quantitative variables, while Spearman correlation coefficient test (r-test) was used to describe the direction of correlation between non-parametric quantitative variables, either positive or negative. P-value shows a statistically significant correlation between studied groups. P<0.05 is considered statistically significant and P<0.001 is considered highly significant.
Results
Demographic and clinical data of SCC and BCC are shown in Table 1.
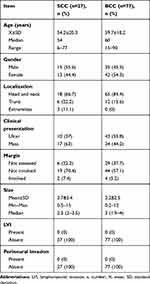 |
Table 1 Demographic and Clinical Data of the SCC and BCC |
Comparison Between Studied Groups Regarding ATG 7 Expressions
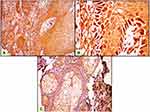
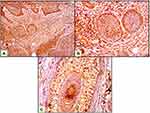
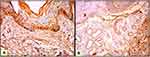
Both epithelial and stromal ATG7 were expressed in all participants, with a significant difference regarding cellular localization. All patients showed a nucleocytoplasmic localization of Atg7 while controls showed both cytoplasmic and nucleocytoplasmic expression (P<0.001) (Figures 1–3). All cases in the control group had belonged to the low H score group, whereas the majority of BCC and SCC had belonged to the high H score group (Table 2).
 |
Table 2 Comparison Between Studied Groups Regarding ATG7 Expressions |
Relation Between ATG 7 H Score and Clinical Parameters of All Patients
No significant association was identified between ATG7 expression and studied clinical parameters of both SCC and BCC patients (Table 3).
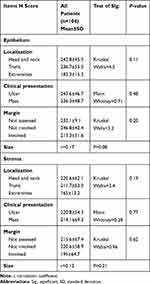 |
Table 3 Relation Between ATG7 H Score and Clinical Parameters of the Patients |
Relationship Between ATG 7 H Score and Clinical Parameters in SCC Patients
No significant association was identified between ATG7 H score and studied clinical parameters of SCC patients (Table 4).
 |
Table 4 Relationship Between ATG7 H Score (Epithelial and Stromal) and Clinical Parameters in SCC Patients |
Relationship Between ATG 7 H Score and Clinical Parameters in BCC Patients
A significant correlation between epithelial H score of Atg7 and size of lesion (P=0.027) was identified. No significant association was found between ATG7 expression and other studied clinical parameters of BCC patients (Table 5).
 |
Table 5 Relationship Between ATG7 H Score (Epithelial and Stromal) and Clinical Parameters in BCC Patients |
Correlation Between H Score (Epithelial and Stromal) in All Patients and BCC and SCC
A significant positive correlation between epithelial and stromal Atg7 H-score in the skin of all patients was identified (r=+0.48, P=0.001). Also a significant positive correlation was identified between epithelial and stromal Atg7 H-score in BCC and SCC (r=+0.29, P=0.009) and (r=+0.61, P=0.001), respectively (Figure 4).
Discussion
Autophagy is a catabolic process in which cell components including proteins, lipids, and entire organelles, are digested using lysosomal enzymes.19 Autophagy is essential for development and maintaining homeostasis in adult organisms. Consequently, an autophagy disorder is connected with multiple pathologies, such as cancer, inflammatory bowel diseases, and neurodegenerative disorders.20
Regarding skin cancer, the importance of autophagy in melanoma has been indicated by numerous studies,21,23 but the role of autophagy in NMSC is still unclear. ATG7 is suggested to be essential for the induction of autophagy.19 The results of the current study revealed that ATG7 was expressed in both patients and controls. However, patients showed significantly higher H-scores than controls besides the predominant nucleocytoplasmic expression which can suggest the role of autophagy in both SCC and BCC.
Similarly, Zhang et al24 found that the phenomena of autophagy occurred in SCC and suggested that 3-methyladenine (3-MA) and 5-fluorouracil (5-FU) combination treatment may be an effective therapy for SCC through modulation of autophagy. In the same point of view, the study of Qiang et al25 showed that autophagy was activated in SCC compared with normal human skin. In addition, Qiang et al proposed that, by suppressing p62-mediated p38 activation, autophagy could encourage cell survival and thus facilitate the growth of tumors under genotoxic stress.
In the same point of view, the role of autophagy was studied in oral squamous cell carcinoma (OSCC). The study of Sakakura et al26 delineated that autophagy elements were related to malignant potential and an unfavorable prognosis in OSCC since autophagy could actively mobilize immune cells toward the cancer bed. Terabe et al27 also suggested that the autophagy markers at the surgical margins could also be an important sign of local recurrence and bad human OSCC prognosis.
Furthermore, the fact that UV radiation exposure is a major risk factor for skin cancer and the suggestion of that UV radiation can induce autophagy in keratinocyte can explain the current results.28 Qiang et al,29 in a mice study, reported that ATG7 promoted UV-induced inflammation and skin tumorigenesis by induction of cytokine expression and secretion, and promotion of prostaglandin 2/Cox-2 expression. In addition, ATG7 deletion in keratinocytes leads to a suppression of stromal microenvironment and tumorigenesis of the epithelium.
When it comes to cellular localization of ATG7, the current study revealed predominant nucleocytoplasmic accumulation in SCC and BCC patients. Luo et al,30 in a trial to explore the role of autophagy in fast-growing tumors, reported that Proteasome 26S Subunit, Non-ATPase 10 (PMD10)/gankyrin promotes autophagy in response to starvation or stress through twocomplementary routes. In cytoplasm, PSMD10 was linked physically to ATG7 and was improved by original nutrient deprivation. PSMD10 was then transported to the nucleus and linked to nuclear heat shock transcription factor 1 (HSF1) onto the ATG7 promoter, upregulated ATG7 expression in the advanced stage of starvation.
Okada et al31 found that immunohistochemical reactivity for ATG7 was detected in the cytoplasm of cellular components in dental follicles and ameloblastomas. There was growing evidence that nucleocytoplasmic shuttling of proteins in the autophagy pathway contributes to the regulation of this process.32 Nucleocytoplasmic localization in this study was in favor of non-melanoma skin cancer could be explained by reluctant autophagy pathway and persistence of inactive form of LC3 in the nucleus.33
Moreover, Huang and Liu33 examined the essential role for nuclear LC3 in starvation-induced autophagy. LC3 is distributed in an acetylated shape in the nucleus as well as in cytoplasm during nutrient rich circumstances. The deprivation of nutrients encourages the transformation of LC3 from the nucleus into the cytoplasm. This shift is dependent on a protein deacetylation. In addition, for LC3 to bind to ATG7 for subsequent lipidation, deacetylation is also necessary. Therefore, the nuclear pool of LC3 is the main source for a membrane conjugated LC3 with a nucleocytoplasmic translocation as a preparation of starved cells for autophagy induction.
Didona et al34 suggest that UV exposure is considered a complete carcinogen as it leads to cellular damage because of the reduction of cell-mediated immune responses, production of reactive oxygen species (ROS) and oxidative damage to DNA. UV exposure affects the p53 expression, which is altered in both AKs and SCC. P53 accumulation can lead to cell cycle arrest or apoptosis. Simon et al,35 upon studying of retrograde signaling from autophagy modulated stress responses, found that ATG7 is an essential protein for the activation of P53-dependent transactivation of P21 leading to cell cycle arrest. ATG7-P53 complexes are found in both the cytoplasm and nucleus of this cells.
Zhu et al36 conducted a study on p62 as a marker for autophagy and compared its expression level in tumor tissues and normal tissues. P62 locates at the autophagosomal membranes and acts as an autophagy receptor through interacting with microtubule-associated protein 1 light chain 3 B (LC3B) and ubiquitinated cargoes. The study demonstrated that p62 protein in tumor cells was elevated and situated primarily in cytoplasm compared to ordinary cells, and this cytoplasmic accumulation predicted bad prognosis in some malignant tumor.
The current study showed that stroma ATG7 expression parallel to the epithelium and both are positively correlated and this means a positive interaction between microenvironment of surrounding tumor and tumor cells. To the best of our knowledge and after a search of English literature, no studies concerning this point were found.
Finally, it is obvious that ATG7 can play a role in pathogenesis of NMSC; however, this study has some limitations in the form of the low number of patients with SCC.
Limitation
Limited number of cases of squamous cell carcinoma.
Conclusion
This study may provide a new insight into the role of autophagy in NMSC pathogenesis and could open the gate for new target therapy for this skin cancer, which needs more study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Duarte AF, Sousa-Pinto B, Haneke E, et al. Risk factors for development of new skin neoplasms in patients with past history of skin cancer: a survival analysis. Sci Rep. 2018;8(1):15744. doi:10.1038/s41598-018-33763-7
2. Rubió-Casadevall J, Hernandez-Pujol AM, Ferreira-Santos MC, et al. Trends in incidence and survival analysis in non-melanoma skin cancer from 1994 to 2012 inGirona, Spain: a population-based study. Cancer Epidemiol. 2016;45:6–10. doi:10.1016/j.canep.2016.09.001
3. Apalla Z, Lallas A, Sotiriou E, et al. Epidemiological trends in skin cancer. Dermatol Pract Concept. 2017;7(2):1–6.
4. Bertozzi N, Simonacci F, Greco MP, et al. Single center evidence for the treatment of basal cell carcinoma of the head and neck. Acta Biomed. 2019;90(1):77–82. doi:10.23750/abm.v90i1.6395
5. Erb P, Ji J, Kump E, et al. Apoptosis and pathogenesis of melanoma and nonmelanoma skin cancer. AdvExp Med Biol. 2008;624:283–95.
6. Yin H, Wu H, Chen Y, et al. The therapeutic and pathogenic role of autophagy in autoimmune diseases. Front Immunol. 2018;9:1512. doi:10.3389/fimmu.2018.01512
7. Yu T, Zuber J, Li J. Targeting autophagy in skin diseases. J Mol Med. 2015;93(1):31–38. doi:10.1007/s00109-014-1225-3
8. Chenog H. Integrating autophagy and metabolism in cancer. Arch Pharm Res. 2015;38(3):358–371. doi:10.1007/s12272-015-0562-2
9. White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15(17):5308–5316. doi:10.1158/1078-0432.CCR-07-5023
10. Guo Y, Zhang X, Wu T, et al. Autophagy in skin diseases. Dermatology. 2019;235(5):380–389. doi:10.1159/000500470
11. Verschooten L, Barrette K, van Kelst S, et al. Autophagy inhibitor chloroquine enhanced the cell death inducing effect of the flavonoid luteolin in metastatic squamous cell carcinoma cells. PLoS ONE. 2012;7(10):1–11. doi:10.1371/journal.pone.0048264
12. Li L, Chen X, Gu H. The signaling involved in autophagy machinery in keratinocytes and therapeutic approaches for skin diseases. Oncotarget. 2016;7:50682–50697. doi:10.18632/oncotarget.9330
13. Lévy J, Romagnolo B. Autophagy, microbiota and intestinal oncogenesis. Oncotarget. 2015;6(33):34067–34068. doi:10.18632/oncotarget.v6i33
14. Xue LY, Chiu SM, Oleinick NL. Atg7 deficiency increases resistance of MCF-7 human breast cancer cells to photodynamic therapy. Autophagy. 2015;6(2):248–255. doi:10.4161/auto.6.2.11077
15. Pascual JC, Belinchón I, Ramos JM. Cutaneous surgery complications in individuals aged 80 and older versus younger than 80 after excision of nonmelanoma skin cancer. J Am Geriatr Soc. 2015;63(1):188–190. doi:10.1111/jgs.13226
16. Nagaral GV. A study on histopathological changes in lesions of vitiligo in Karnataka population. Int J Res Dermatol. 2017;3(1):94. doi:10.18203/issn.2455-4529.IntJResDermatol20170794
17. Sharifi MN, Mowers EE, Drake LE, et al. Measuring autophagy in stressed cells. In: Methods in Molecular Biology. Vol. 1292. Clifton, NJ; 2015:129–150
18. Bilalovic N, Sandstad B, Golouh R, et al. CD10 protein expression in tumor and stromal cells of malignant melanoma is associated with tumor progression. Mod Pathol. 2004;17(10):1251–1258. doi:10.1038/modpathol.3800174
19. Arakawa S, Honda S, Yamaguchi H, et al. Molecular mechanisms and physiological roles of Atg5/Atg7-independent alternative autophagy. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(6):378–385.
20. Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12(9):823–830. doi:10.1038/ncb0910-823
21. Liu H, He Z, Simon H-U. Targeting autophagy as a potential therapeutic approach for melanoma therapy. Semin Cancer Biol. 2013;23(5):352–360. doi:10.1016/j.semcancer.2013.06.008
22. Maes H, Agostinis P. Autophagy and mitophagy interplay in melanoma progression. Mitochondrion. 2014;19:58–68. doi:10.1016/j.mito.2014.07.003
23. Ndoye A, Weeraratna AT. Autophagy- an emerging target for melanoma therapy. F1000Res. 2016;5:1888. doi:10.12688/f1000research
24. Zhang L, Zhang J, Chen L, et al. Autophagy in human skin squamous cell carcinoma: inhibition by 3-MA enhances the effect of 5-FU-induced chemotherapy sensitivity. Oncol Rep. 2015;34:3147–3155. doi:10.3892/or.2015.4302
25. Qiang L, Wu C, Ming M, et al. Autophagy controls p38 activation to promote cell survival under genotoxic stress. J Biol Chem. 2013;288:1603–1611. doi:10.1074/jbc.M112.415224
26. Sakakura K, Takahashi H, Kaira K, et al. Immunological significance of the accumulation of autophagy components in oral squamous cell carcinoma. Cancer Sci. 2015;106:1–8. doi:10.1111/cas.2015.106.issue-1
27. Terabe T, Uchida F, Nagai H, et al. Expression of autophagy-related markers at the surgical margin of oral squamous cell carcinoma correlates with poor prognosis and tumor recurrence. Hum Pathol. 2018;73:156–163. doi:10.1016/j.humpath.2017.11.019
28. Sample A, He -Y-Y. Autophagy in UV damage response. Photochem Photobiol. 2017;93(4):943–955.
29. Qiang L, Sample A, Shea CR, et al. Autophagy gene ATG7 regulates ultraviolet radiation-induced inflammation and skin tumorigenesis. Autophagy. 2017;13:2086–2103. doi:10.1080/15548627.2017.1380757
30. Luo T, Fu J, Xu A, et al. PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy. 2016;12(8):1355–1371. doi:10.1080/15548627.2015.1034405
31. Okada M, Oikawa M, Miki Y, et al. Immunohistochemical assessment of ATG7, LC3, and p62 in ameloblastomas. J Oral Pathol Med. 2014;43:606–612. doi:10.1111/jop.2014.43.issue-8
32. Liang XH, Yu J, Brown K, et al. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449.
33. Huang R, Liu W. Identifying an essential role of nuclear LC3 for autophagy. Autophagy. 2015;11:852–853. doi:10.1080/15548627.2015.1038016
34. Didona D, Paolino G, Bottoni U, et al. Non melanoma skin cancer pathogenesis overview. Biomedicines. 2018;6(1):2. doi:10.3390/biomedicines6010006
35. Simon H, Friis R, Tait SWG, et al. Retrograde signaling from autophagy modulates stress responses. Sci Signal. 2017;28(10):468–478.
36. Zhu L, Wang Y, He J, et al. Cytoplasmic SQSTM1/P62 accumulation predicates a poor prognosis in patients with malignant tumor. J Cancer. 2018;9(21):4072–4086. doi:10.7150/jca.26399
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.